Transcription
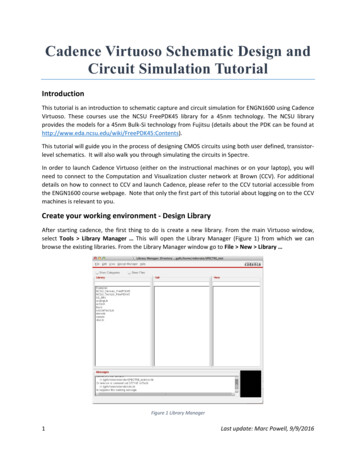
Automated library preparationof small genomes using DNApreparation kitsWritten byMatthew Akana, M.Sc., Kinnari Watson, PhDFor the Illumina DNA Prep kit, the DNA was tagmentedABSTRACTand tagged using Dual Index adapters on the OT-2.Three library prep kits – Illumina DNA Prep Kit, KAPADNA Prep for lambda DNA samples (n 8, 100 ng) wasHyperPrep kit, and NEBNext Ultra II Kit – werequantified, normalized to a 4 nM library, pooled intoautomated with the Opentrons OT-2 for fast and robusta single multiplexed sample, and then ran on a 2x75preparation of high quality libraries for next generationflow cell on Illumina MiSeq for genome sequencing. Thesequencing.workflow included generating DNA prep libraries for Libraries prepared with 100 ng input were preparedLambda (n 24) and Human Genomic DNA samples (n 8). from lambda and human genomic DNA. Comparable performance was observed for librariesFor both HyperPrep and NEBNext Ultra II DNA prep kits,prepared with the three kits on the OT-2 in terms ofDNA libraries were constructed for fragmented lambdasample variability and yield. Similar sequencing metricsDNA (n 8, 100 ng) on an OT-2. Barcodes used were fromsuch as barcode balance and sequence alignmentKAPA Unique Dual Index Adapters for HyperPrep, andcoverage wereNEB Multiplex Dual Index Primers Set 1 for NEBNext Ultraalso observed.II. Samples were quantified, normalized, and pooled intoa 4 nM library and sequenced on a MiSeq with a 2x75flow cell. The workflow included generating libraries forINTRODUCTIONlambda fragmented DNA (n 24) and human genomicDNA library preparation is a critical step for next-DNA (n 8).generation sequencing (NGS). Automating DNAlibrary preparation provides a streamlined approachThe sequencing data were demultiplexed by Illumina’sfor constructing high-yield libraries for sequencingBaseSpace according to the adapter barcode sequencewhile minimizing hands-on time. Here, we describeand aligned to the reference genome. The coverage mapthe efficiency of automating the Illumina DNA Prepof the genome was analyzed using Geneious Primekit (Illumina, Cat. No. 20018705), KAPA HyperPrep kit2021.2.21. Our data showed low sample variability and(Roche , Cat. No. 7962363001), and NEBNext Ultra II kitreliable uniformity for libraries prepared using DNA prep(New England Biolabs , Cat. No. E7645L) on the OT-2kits on the OT-2. robotic liquid handling platform.SCHEMATIC OF THE OT-2 WORKFLOW PROTOCOLMATERIALS AND METHODSFor the Illumina DNA Prep kit, the DNA and IDT forOverview of the Illumina DNA Prep kit, KAPA HyperPrep Kit,Illumina DNA/RNA UD Indexes Set A (Illumina, Cat. No.and NEBNext Ultra II and the Opentrons OT-2 Platform20027213) were tagmented on the OT-2 (Figure 1). Awash step is necessary for removing residual DNA andIllumina has a patented NGS tagmentation workflow thatadapters. Next, PCR amplification was conducted using anutilizes bead-linked transposomes, which differs fromOT-2 on-deck thermocycler with a programmable lid andthe KAPA HyperPrep and NEBNext Ultra II workflows-inblock temperatures.solution process for DNA end-repair and A-tailing.
For both HyperPrep and NEBNext Ultra II DNA prep kits,DNA amplification was performed using an on-deckfragmentation was performed with NEB Fragmentasethermocycler on the OT-2.for 16 minutes for lambda and 20 minutes for humangenomic DNA, followed by a cleanup step with AMPure THE WORKFLOW LAYOUTXP Beads (Beckman Coulter, Cat. No. A63881). DNA endrepair/A tailing, followed by the AMPure XP beads cleanupThe layout of the Opentrons OT-2 platform includes thestep, barcode, or adapter ligation, was performed on themodules, labware, and DNA Prep reagents (Figure 2).OT-2 (Figure 1). Barcodes used were from KAPA UniqueAlthough the workflow consists of automated pipettingDual Index Adapters (Roche, Cat. No. 08861919702)on the OT-2, manual intervention is required to move thefor HyperPrep and NEB Multiplex Dual Index Primerssample plate between the on-deck Thermocycler and theSet 1 (NEB, Cat. No. E7335L) for NEBNext Ultra II.Magnetic Block and reset the pipette tip racks duringHyperPrepNEBNext Ultra II100ng DNA100ng Fragmented DNA100ng Fragmented DNATagmentationEnd Repair / A TailingEnd Repair / A TailingTagmentation WashBarcode LigationAdapter LigationAmplificationAMPure CleanupOT-2 ProtocolIllumina DNA PrepOT-2 ProtocolOT-2 Protocolthe protocol.AMPure CleanupAmplificationAMPure CleanupBarcode AmplificationAMPure CleanupAMPure CleanupQuantify, Normalize, PoolQuantify, Normalize, PoolDenature, SequenceDenature, SequenceQuantify, Normalize, PoolDenature, SequenceFigure 1: NGS Workflows with OT-2. The blue-shaded boxes indicate the steps of the DNA Prep workflow automated on the OT-2Figure 2: Opentrons OT-2 deck layout equipped with1modules, labware, and DNA Prep reagents. Thisstandardized deck layout is shared with HyperPrep andNEBNext Ultra II. This workflow has all pipetting automatedon the OT-2 and requires manual intervention to move thesample plate (1) between the on-deck Thermocycler andMagnetic Block (2), as well as resetting tip racks during therun. The deck layout includes 1 x NEST 12 well reservoir,1 x 96 well aluminum block, 1 x Eppendorf 200 µl PCR Plateon the Thermocycler, and a 1 x NEST 0.1mL PCR plate on32the Temperature module (3). Modules include a MagneticModule, Temperature module, Thermocycler module, andP20 and P300 8-Channel pipettes.
fragment sizes of the DNA prep libraries to determineRESULTSthe reliable uniformity for libraries prepared on the OT-2DNA Library Amplification(Table 3A–3C). Similarly, consistent sequencing coverageDNA libraries were measured using PicoGreen on aof the 48 kb lambda genome showed uniformity acrossQubit 4 Fluorometer to determine concentration. The CV8-plexed DNA prep libraries constructed on the OT-2(coefficient of variation is the standard deviation divided(Table 4). by average). The final elution volume for all the librarieswas 30 µL. For libraries prepared using the IlluminaBarcode balance was accurate for the DNA Prep samplesDNA Prep showed a CV of 10.7% for (n 8, 100 ng) and ashowing an average of 11.2%–12.31% for IlluminaCV of 16.6% for (n 24, 100 ng) (Table 1A). For librariesDNA Prep, 10.6%–14.1% for the KAPA HyperPrep, andprepared using the HyperPrep, showed a CV of 13.4% for10.5%–14.6% for the NEBNext Ultra II (Figure 3). The(n 8, 100 ng) and a CV of 17.6 for (n 24, 100 ng) (Tablecomparisons of the DNA prep kits allocation of tips and1B). Libraries prepared using the NEBNext Ultra II showedthe duration of OT-2 protocol (Table 5).a CV of 12.1% for (n 8) and a CV of 14.5% for (n 24)(Table 1C). Comparisons for the yield (ng/µl), CV (%), andCONCLUSIONfragment size (bp) for lambda and human DNA librariesprepared on the OT-2 are shown in Table 2.We validated Illumina DNA Prep, KAPA HyperPrep,and NEBNext Ultra II on the Opentrons OT-2 andSmall Genome Sequencing of DNA Prep Librariesdemonstrated its use for library preparation for nextThe multiplexed lambda DNA prep libraries (n 8, 100generation sequencing. We showed that the DNA Prepng) were sequenced using a 2x75 flow cell on a Illuminasamples displayed low variability across replicates andMiSeq instrument. The second batch of DNA prep librarieschemistries, even sample barcode balance, and high(n 24, 100 ng) were processed in 3 separate columns oncoverage of the sequence alignments, demonstrating thatthe OT-2 to account for any inter-column variability withinthe OT-2 can be used to reliably automate preparation ofa batch. The Agilent 5300 Fragment Analyzer calculatedhigh-quality DNA libraries.ILLUMINA DNA PREP ON 100 NG LAMBDA8x Replicates24x ReplicatesColumn 1Column 1Column 2Column Table 1A: Low variability among libraries constructed on the Opentrons OT-2.Final eluted volume is 30 µl, values are ng/µl. This table shows the final yield of each sample after Illumina DNA Prep.
HYPERPREP (NG/µL)8x Replicates24x ReplicatesColumn 1AverageColumn 1Column 2Column 7.6%Average15.9CVCV13.4%Table 1B: Low variability among libraries constructed on the Opentrons OT-2.Final eluted volume is 30 µl, values are ng/µl. This table shows the final yield of each sample after HyperPrep.NEBNEXT ULTRA II (NG/µL)8x Replicates24x ReplicatesColumn 1Column 1Column 2Column Table 1C: Low variability among libraries constructed on the Opentrons OT-2.Final eluted volume is 30 µl, values are ng/µl. This table shows the final yield of each sample after NEBNext Ultra II.SUMMARYINPUTSAMPLE TYPESAMPLESYIELD (NG/µL)CVFRAGMENT SIZE (BP)Illumina DNA Prep100 ngIllumina DNA Prep100 ngLambda828.2310.7%310 15Lambda2435.5615.9%Illumina DNA Prep316 15100 ngHuman Genomic2410.6917.3%247 15HyperPrep100 ng FraggedLambda815.9413.4%280 10HyperPrep100 ng FraggedLambda249.1814.6%280 10HyperPrep100 ng FraggedHuman Genomic816.6111.4%283 10NEB Ultra II100 ng FraggedLambda818.6112.1%259 12NEB Ultra II100 ng FraggedLambda2427.1014.0%255 12NEB Ultra II100 ng FraggedHuman Genomic819.4314.8%243 12Table 2: Comparison of NGS Sample Library Preps on the OT-2.
Table 3A: Illumina DNA Prep Sample Fragment SizesTable 3B: HyperPrep Sample Fragment SizesTable 3C: NEBNext Ultra II Sample Fragment SizesAverageILLUMINA DNA PREPHYPERPREPNEBNEXT ULTRA II7865x6883x7589xTable 4: NGS Sample Prep sequencing coverage. A comparison of Lambda Genome coverage of 2x75 Miseq runsof 8x Multiplexed sample libraries.
Illumina DNA Prep (%)Roche HyperPrep (%)NEBNext Ultra (%)Sample 114.311.311.9Sample 216.111.412.6Sample 310.413.612.1Sample 414.411.814.6Sample 59.313.514.8Sample 612.714.110.4Sample 711.710.610.5Sample 811.113.813.0Figure 3: Uniform sample barcode representation. This chart demonstrates the even sample barcode balance within the sequencing run byoverall read percentage.TIP BOXES REQUIRED(WITHOUT REUSING TIPS)TIP BOXES REQUIRED(WITH REUSING TIPS)p20p300p20p300Time24x15132 Hrs 44 Min16x13122 Hrs 14 Min8x12111 Hrs 48 Min24x15133 Hrs 9 Min16x14122 Hrs 42 Min8x12112 Hrs 20 Min24x25133 Hrs 9 Min16x14122 Hrs 42 Min8x12112 Hrs 20 MinIllumina DNA PrepHyperPrepNEBNext Ultra IITable 5: Comparison of NGS library prep configuration experiment duration and tip usage.REFERENCES1.Geneious Prime 2021.2.2
the KAPA HyperPrep and NEBNext Ultra II workflows-in solution process for DNA end-repair and A-tailing. For the Illumina DNA Prep kit, the DNA was tagmented and tagged using Dual Index adapters on the OT-2. DNA Prep for lambda DNA samples (n 8, 100 ng) was quantified, normalized to a 4 nM library, pooled into