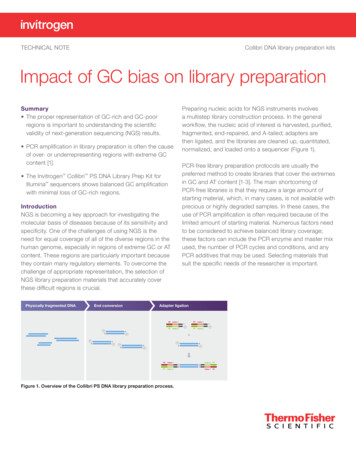
Transcription
Technical Data SheetKAPA LTP Library Preparation KitIllumina platformsKit Codes and ComponentsKR0453 – v3.13This Technical Data Sheet provides product informationand a detailed protocol for the KAPA LTP LibraryPreparation Kit (Illumina platforms), product codesKK8230, KK8231, KK8232 and KK8233.ContentsKK8230KK823110 librariesProduct Description .2KK8232KK8233Product Applications .250 librariesProduct Specifications .2Shipping and storage .2Handling .2Quality control .2Important Parameters .3Reaction setup .3Safe stopping points .3Paramagnetic SPRI beads and reaction cleanups .3Input DNA and fragmentation .4Cleanups after end repair and A-tailing .4Adapter design and concentration .4Post-ligation cleanup .5Size selection .5Library amplification .6Evaluating the success of library construction .7Process Workflow .8Library Construction Protocol .9Appendix 1:Library construction guidelines for target enrichment withthe Roche Nimblegen SeqCap EZ system .16KAPA End Repair Buffer (10X)KAPA End Repair Enzyme MixKAPA A-Tailing Buffer (10X)KAPA A-Tailing EnzymeKAPA Ligation Buffer (5X)KAPA DNA LigaseKAPA PEG/NaCl SPRI SolutionKAPA HiFi HotStart ReadyMix (2X)*100 µl50 µl50 µl30 µl100 µl50 µl5 ml250 µlKAPA End Repair Buffer (10X)500 µlKAPA End Repair Enzyme Mix250 µlKAPA A-Tailing Buffer (10X)250 µlKAPA A-Tailing Enzyme150 µlKAPA Ligation Buffer (5X)500 µlKAPA DNA Ligase250 µlKAPA PEG/NaCl SPRI Solution20 mlKAPA HiFi HotStart ReadyMix (2X)* 1.25 ml*KK8231 and KK8233 are available for PCR-free workflows and do notcontain KAPA HiFi HotStart ReadyMix for library amplification.Quick Notes The protocol provided in this document is ageneric prototype, and may require additionaltailoring and optimization. The process workflow (p. 8) provides an overviewof the library construction process and optionsfor size selection. The KAPA NGS Library Preparation TechnicalGuide contains more detailed informationabout library construction parameters, and mayfacilitate protocol development and optimization. Separate, concentrated enzyme formulationsand reaction buffers for end repair, A-tailing, andligation provide the best combination of productstability, convenience, and efficiency. Adapters and PCR primers are not supplied withthis kit, and can be obtained from any reputableoligonucleotide vendor. SPRI beads are not included in the kit, but thePEG/NaCl SPRI Solution required for "withbead" reaction cleanups is provided.Effective date: July 20131
KAPA LTP Library Preparation KitIllumina platformsTechnical Data SheetProduct DescriptionProduct ApplicationsThe KAPA LTP Library Preparation Kit is designed for theconstruction of libraries for Illumina sequencing, startingfrom fragmented, double-stranded DNA. The kit providesall of the enzymes and reaction buffers required for thefollowing steps of library construction:The KAPA LTP Library Preparation Kit is ideally suitedfor low-throughput, manual NGS library constructionworkflows that involve end repair, A-tailing, adapterligation, and library amplification (optional). The protocolmay be adapted for incorporation into workflows for awide range of NGS applications, including:1. End repair, which produces blunt-ended, 5'-phosphorylated fragments.2. A-tailing, during which dAMP is added to the 3'-endsof blunt-ended dsDNA library fragments.3. Adapter ligation, during which dsDNA adapters with3'-dTMP overhangs are ligated to 3'-A-tailed libraryfragments.4. Library amplification (optional), which employs PCRto amplify library fragments carrying appropriateadapter sequences on both ends.The kit has been validated for library construction from100 ng – 5 µg of human genomic DNA for whole-genomeshotgun sequencing or targeted sequencing by solutionhybrid selection (capture). For smaller genomes, or lowercomplexity samples, such as ChIP DNA, amplicons, orcDNA (for RNA-seq), successful library construction hasbeen achieved from low nanogram to picogram quantities( 100 pg) of input DNA.The kit provides all of the enzymes and buffers requiredfor library construction and amplification, but does notinclude adapters, PCR primers, or SPRI beads. Enzymeformulations and reaction buffers for end repair, A-tailing,and ligation are supplied in convenient, concentratedformats.Efficient, cost-effective reaction cleanups and higherrecovery of input DNA are achieved throughimplementation of the "with-bead" strategy developedat The Broad Institute of MIT & Harvard and FoundationMedicine1. The kit includes PEG/NaCl SPRI (Solid PhaseReversible Immobilization) Solution for this purpose.In order to maximize sequence coverage uniformity, itis critical to minimize library amplification bias. KAPAHiFi DNA Polymerase has been designed for low-bias,high-fidelity PCR, and is the reagent of choice for NGSlibrary amplification2, 3, 4. KAPA LTP Library PreparationKits (KK8230 and KK8232) include KAPA HiFi HotStartReadyMix (2X), a ready-to-use PCR mix comprising all thecomponents for library amplification, except primers andtemplate. Kits without an amplification module (KK8231and KK8233) are available for PCR-free workflows.These kits can also be combined with KAPA HiFi RealTime Library Amplification Kits (KK2701 and KK2702),or with KAPA HiFi HotStart Uracil ReadyMix (KK2801and KK2802) for the amplification of libraries that haveundergone bisulfite-treatment.1.2.3.4.2Fisher, S. et al. Genome Biology 12, R1 (2011).Oyola, S.O. et al. BMC Genomics 13, 1 (2012).Quail M.A. et al. Nature Methods 9, 10 – 11 (2012).Quail M.A. et al. BMC Genomics 13: 341 (2012). Whole-genome shotgun sequencing Targeted sequencing by solution hybrid selection(i.e. exome or custom capture using the RocheNimblegen , Agilent SureSelect, Illumina TruSeq , orIDT xGen Lockdown Probes systems) ChIP-seq RNA-seq Methyl-seq (in combination with the KAPA HiFiHotStart Uracil ReadyMix)Specific guidelines for the construction of libraries fortarget enrichment using the Roche Nimblegen SeqCapEZ system may be found in Appendix 1.Product SpecificationsShipping and storageThe enzymes provided in this kit are temperature sensitive,and appropriate care should be taken during shipping andstorage. KAPA Library Preparation Kits are shipped on dryice or ice packs, depending on the country of destination.Upon receipt, immediately store enzymes and reactionbuffers at -20 C in a constant-temperature freezer. ThePEG/NaCl SPRI Solution should be protected from light,and stored at -20 C. For short-term use, the PEG/NaClSPRI Solution may be stored at 4 C for up to 2 months.When stored under these conditions and handledcorrectly, the kit components will retain full activity untilthe expiry date indicated on the kit label.HandlingAlways ensure that components have been fully thawedand thoroughly mixed before use. Keep all enzymecomponents and master mixes on ice as far as possibleduring handling and preparation. KAPA HiFi HotStartReadyMix (2X) contains isostabilizers and may not freezecompletely, even when stored at -20 C. Nevertheless,always ensure that the KAPA HiFi HotStart ReadyMix isfully thawed and thoroughly mixed before use. PEG/NaClSPRI Solution does not freeze at -20 C, but should beequilibrated to room temperature and thoroughly mixedbefore use.Quality controlAll kit components are subjected to stringent functionalquality control, are free of detectable contaminating exoand endonuclease activities, and meet strict requirementswith respect to DNA contamination. Please contactsupport@kapabiosystems.com for more information.
KAPA LTP Library Preparation KitIllumina platformsImportant ParametersLibrary construction workflows must be tailored andoptimized to accommodate specific experimental designs,sample characteristics, sequencing applications, andequipment. The protocol provided in this document is ageneric prototype, and there are many parameters whichmay be adjusted to optimize performance, efficiency, andcost-effectiveness.In addition to the information in this section, pleaseconsult the KAPA NGS Library Preparation TechnicalGuide and/or contact support@kapabiosystems.comfor further guidelines when designing or optimizing yourlibrary construction workflow.Reaction setupWhile this kit is intended for low-throughput, manuallibrary construction, the protocol is designed to beautomation-friendly in order to facilitate the transition toautomation should throughput requirements grow overtime. For this reason, and to enable a streamlined “withbead” strategy, reaction components are combined intomaster mixes, rather than dispensed separately intoindividual reactions. When processing multiple samples,prepare 5 – 10% excess of each master mix, to allow forsmall inaccuracies during dispensing.Libraries may be prepared in standard reaction vesselsincluding 1.5 ml microtubes, PCR tubes, strip tubes,or PCR plates. Always use plastics that are certified tobe free of DNAses, RNAses, and nucleases. Low DNAbinding plastics are recommended. When selecting themost appropriate plasticware for your workflow, considercompatibility with: the magnet used during SPRI bead manipulations. vortex mixers and centrifuges, where appropriate. heating blocks or thermocyclers used for reactionincubations and/or library amplification.Safe stopping pointsThe library construction process, from end repair tofinal, amplified library, can be performed in 4 – 8 hours,depending on the specific workflow, number of samplesbeing processed, and experience. If necessary, theprotocol may be paused safely after any of the beadcleanup steps, as described below: After the end repair cleanup (Steps 3.1 – 3.13),resuspend the washed beads in 20 µl of 1X A-TailingBuffer (without enzyme; Step 4.1), and store thereactions at 4 C for up to 24 hours. After the A-tailing cleanup (Steps 5.1 – 5.13), resuspendthe washed beads in 20 µl of 1X Ligation Buffer (withoutenzyme or adapter; Step 6.1), and store the reactionsat 4 C for up to 24 hours.Technical Data Sheet After the first post-ligation cleanup (Steps 7.1 – 7.13),resuspend the washed beads in the appropriatevolume of 10 mM Tris-HCl (pH 8.0) as outlined in Step7.14, and store the reactions at 4 C for up to 24 hours.DNA solutions containing beads must not be frozen,and beads must not be stored dry, as this is likely todamage the beads and result in sample loss. To resumethe library construction process, centrifuge the reactionvessels briefly to recover any condensate, and add theremaining components required for the next enzymaticreaction in the protocol (see Tables 4B and 5B onp. 10). If the protocol was paused after the first postligation cleanup, continue directly with the second postligation cleanup (Step 7.16), dual-SPRI size selection(Step 8.1), or size selection using an alternative method.Adapter-ligated DNA that has been completely cleaned upor size-selected can be stored at 4 C for one week, or at-20 C for at least one month before amplification, targetenrichment, and/or sequencing. To avoid degradation,always store DNA in a buffered solution (10 mM Tris-HCl,pH 8.0) and minimize the number of freeze-thaw cycles.Paramagnetic SPRI beads and reaction cleanups Cleanups should be performed in a timely manner toensure that enzyme reactions do not proceed beyondoptimal incubation times. This protocol has been validated using Agencourt AMPure XP reagent (Beckman Coulter, part numberA63880, A63881, or A63882). Solutions and conditionsfor DNA binding and size selection may differ if otherbeads are used. Observe all the manufacturer's storage and handlingrecommendations for AMPure XP reagent. Beads will settle gradually; always ensure that theyare fully resuspended before aspirating AMPure XPreagent. The incubation times provided for reaction cleanupsand size selection are guidelines only, and shouldbe modified/optimized according to your currentprotocols, previous experience, and specific equipmentand samples in order to maximize library constructionefficiency and throughput. The time required to completely capture magneticbeads varies according to the reaction vessel andmagnet used. It is important to not discard or transferany beads with the removal or transfer of supernatant.Capture times should be optimized accordingly. The volumes of 80% ethanol used for bead washesmay be adjusted to accommodate smaller reactionvessels and/or limited pipetting capacity, but it isimportant that the beads are entirely submerged duringthe wash steps. Where possible, use a wash volumethat is equal to the volume of sample plus AMPure XPreagent or PEG/NaCl SPRI Solution.3
KAPA LTP Library Preparation KitIllumina platformsParamagnetic SPRI beads and reaction cleanups(continued) It is important to remove all ethanol before proceedingwith subsequent reactions. However, over-drying ofbeads may make them difficult to resuspend, andmay result in a dramatic loss of DNA. With optimizedpipetting, drying of beads for 3 – 5 min at roomtemperature should be sufficient. Drying of beads at37 C is not recommended. Where appropriate, DNA should be eluted from beadsin elution buffer (10 mM Tris-HCl, pH 8.0). Elution ofDNA in PCR-grade water is not recommended, asDNA is unstable in unbuffered solutions.Input DNA and fragmentation This protocol has been validated for library constructionfrom 100 ng – 5 µg of appropriately fragmented,double-stranded DNA. However, libraries can beprepared from lower input amounts if the samplerepresents sufficient copies to ensure the requisitecoverage and complexity in the final library. Successfullibrary construction has been achieved from 100 pgof ChIP DNA, low nanogram quantities of cDNA ormicrobial DNA, and 1 – 10 ng of high-quality human ormouse genomic DNA.Technical Data Sheetin which case the end repair reaction setup shouldbe adjusted accordingly. Please contact support@kapabiosystems.com for more information.Cleanups after end repair and A-tailing This protocol provides for 1.7X – 1.8X cleanups afterend repair and A-tailing. This ratio of PEG/NaCl SPRI Solution to sample volume will retain most DNAfragments larger than 75 bp. If you wish to retain verysmall DNA fragments, the PEG/NaCl SPRI Solutionto sample ratio can be increased to 2X – 3X for allcleanups prior to adapter ligation. When performing library construction in standardPCR plates, a 3X cleanup after end repair is notpossible, as the maximum working volume per wellis usually 200 µl. To achieve a 3X cleanup after endrepair, libraries should be prepared in 500 µl PCRtubes or 1.5 ml microtubes, or the end repair reactionshould be scaled down. Please contact support@kapabiosystems.com for more information.Adapter design and concentration This protocol has been validated using standard,indexed Illumina TruSeq "forked" adapters, but thekit is compatible with other adapters of similar design. The above typically refers to the input into the endrepair reaction. If input DNA is quantified beforefragmentation, and/or fragmented DNA is subjected tocleanup or size selection prior to end repair, the actualinput into library construction may be significantlylower. This should be taken into account whenevaluating the efficiency of the process and/or duringoptimization of library amplification cycle number. Adapter concentration affects ligation efficiency, aswell as adapter and adapter-dimer carry-over in postligation cleanups. The optimal adapter concentrationfor your workflow represents a compromise betweencost and the above factors. Your choice of postligation cleanup and size-selection options should beinformed by your choice of adapter concentration.Please refer to the next subsection for more details. The proportion of fragmented DNA that is successfullyconverted to adapter-ligated molecules decreases asinput is reduced. When starting library construction(end repair) with 100 ng fragmented DNA, 15 – 40%of input DNA is typically recovered as adapter-ligatedmolecules, whereas the recovery typically rangesfrom 0.5 to 15% for libraries constructed from 100 pg– 100 ng DNA. These figures apply to high qualityDNA and can be significantly lower for DNA of lowerquality, e.g. FFPE samples. Workflows that containadditional SPRI cleanups or size selection prior tolibrary amplification are likely to result in a lower yieldof adapter-ligated molecules. Ligation efficiency is robust for adapter:insertmolar ratios ranging from 10:1 to 50:1. As a generalguideline, we recommend an adapter:insert molarratio of 10:1, for libraries constructed from 100 ngfragmented DNA. This translates to different finaladapter concentrations for libraries with differentsize distributions (see Table 1 on the next page). Anadapter:insert molar ratio 10:1 may be beneficialfor libraries constructed from lower amounts of inputDNA. Solutions containing high concentrations of EDTA andstrong buffers may negatively affect the end repairreaction, and should be avoided. If fragmented DNAwill not be processed (i.e. subjected to cleanup or sizeselection) prior to end repair, DNA should be fragmentedin 10 mM Tris-HCl (pH 8.0 or 8.5) with 0.1 mM EDTA.Fragmentation in water is not recommended. In some circumstances it may be convenient tofragment input DNA in 1X KAPA End Repair Buffer,4 While it is not necessary to adjust adapterconcentrations to accommodate moderate sampleto-sample variations, we recommend using an adapterconcentration that is appropriate for the range of inputDNA concentrations. The best way to accommodate different adapterconcentrations within a batch of samples processedtogether, is to vary the concentration of adapterstock solutions, and dispense a fixed volume (5 µl) ofeach adapter. The alternative – using a single stocksolution, and dispensing variable volumes of adapterinto ligation reactions – is less compatible with higherthroughput or automated workflows.
KAPA LTP Library Preparation KitIllumina platformsTechnical Data SheetTable 1. Recommended adapter concentrations.Recommended adapter concentration for DNA sheared to an average size ofInsert DNA per 50 µlend repair reaction175 bp350 bp500 bpStockFinalStockFinalStockFinal3 – 5 µg60 µM6 µM30 µM3 µM21 µM2.1 µM1 µg20 µM2 µM10 µM1 µM7 µM0.7 µM500 ng10 µM1 µM5 µM500 nM3.5 µM350 nM100 ng2 µM200 nM1 µM100 nM700 nM70 nM10 ng200 nM20 nM100 nM10 nM70 nM7 nMPost-ligation cleanupSize selection It is important to remove excess unligated adapterand adapter-dimer molecules from the library prior tolibrary amplification or cluster generation. Size selection requirements vary widely accordingto specific applications. Depending on preference,the dual-SPRI size selection procedures presentedin this protocol may be omitted entirely, modified, orreplaced with alternative size selection procedures.Size selection may be carried out at alternative pointsin the overall workflow, for example: While a single SPRI bead cleanup removes mostunligated adapter and adapter-dimer, a secondSPRI bead cleanup is recommended to eliminateany remaining adapter species from the library.The amount of adapter and adapter-dimer carriedthrough the first cleanup is dependent on the adapterconcentration in the ligation reaction. If size selection is carried out between adapter ligationand library amplification (or clustering), a single postligation cleanup with SPRI beads (1X) is usuallysufficient prior to size selection. If no post-ligation sizeselection is carried out, two consecutive 1X SPRI bead cleanups are recommended. The volume in which washed beads are resuspendedafter the post-ligation cleanup(s) should be adjusted tosuit your chosen workflow: If proceeding directly to library amplification,determine an appropriate final volume in which toelute the library DNA, keeping in mind that you maywish to divert and/or reserve some of this librarymaterial for archiving and/or QC purposes. Sincean optimized 50 µl library amplification reactionshould yield 1 µg of DNA, and can accommodatea maximum of 20 µl template DNA, an elutionvolume of 22 – 32 µl is recommended. If proceeding with size selection, elute the libraryDNA in an appropriate volume according to the sizeselection method of choice. For the dual-SPRI sizeselection procedure described here, beads have tobe resuspended in a final elution volume of 100 µl. prior to end repair of fragmented DNA. immediately before library amplification (as outlinedin the protocol that follows). after library amplification. Size selection inevitably leads to a loss of samplematerial. Depending on the details, these losses canbe dramatic ( 80%), and may significantly increasethe number of amplification cycles needed to generatesufficient material for the next step in the process(capture or sequencing). The potential advantages ofone or more size selection steps in a library constructionworkflow should be weighed against the potential lossof library complexity, especially when input DNA islimited. A carefully optimized fragmentation protocol,especially for shorter insert libraries and/or readlengths, may eliminate the need for size selection,thereby simplifying the library construction processand limiting sample losses. Over-amplification of libraries often results inthe observation of secondary, higher molecularweight peaks when amplified libraries are analyzedelectrophoretically. These higher molecular weightpeaks are artefacts, and typically contain authenticlibrary molecules of the appropriate length. To eliminatethese artefacts, optimization of library amplificationreaction parameters (cycle number and/or primerconcentration), rather than post-amplification sizeselection, is recommended. See Library amplificationon pp. 6 – 7 for more information.5
KAPA LTP Library Preparation KitIllumina platformsSize selection (continued) KAPA Ligation Buffer contains high concentrations ofPEG 6000, which will interfere with efficient dual-SPRI size selection and can affect the efficiency of othersize selection techniques if not removed. It is thereforeimportant to perform at least one post-ligation SPRI bead cleanup (1X) prior to dual-SPRI or any other sizeselection method. The dual-SPRI size selection procedure described inSection 8 of the protocol is designed for selection ofadapter-ligated fragments approximately 250 – 450 bpin size. Consult the KAPA NGS Library PreparationTechnical Guide if you wish to select a different rangeof fragment sizes. "Forked" adapters with long single-stranded arms(such as the TruSeq design) noticeably affect thesize-dependent binding of DNA to SPRI beads, aswell as the apparent size of fragments determinedby some electrophoresis instruments (e.g. thoseemploying microfluidic chips and tapes). Sizeselection parameters will therefore require optimizationdepending on a number of factors, including: The design of the adapters used. The point at which size selection is applied in theprotocol; size selection of dsDNA fragments priorto library construction or after amplification mayrequire different parameters than those used forpost-ligation size selection of fragments carryingforked adapter ends. Dual-SPRI size selection is sensitive to multiplefactors that are beyond the scope of this document.The KAPA NGS Library Preparation Technical Guidecontains additional guidelines for the optimization ofdual-SPRI size selection parameters. Any dual-SPRI size selection protocol should be carefully optimizedand validated before it is used for precious samples.Library amplification KAPA HiFi HotStart, the enzyme provided in theKAPA HiFi HotStart ReadyMix, is an antibodybased hot start formulation of KAPA HiFi DNAPolymerase, a novel B-family DNA polymeraseengineered for increased processivity and high fidelity.KAPA HiFi HotStart DNA Polymerase has 5'g3'polymerase and 3'g5' exonuclease (proofreading)activities, but no 5'g3' exonuclease activity.The strong 3'g5' exonuclease activity results insuperior accuracy during DNA amplification. Theerror rate of KAPA HiFi HotStart DNA Polymerase is2.8 x 10-7 errors/base, equivalent to 1 error in 3.5 x 106nucleotides incorporated. In library amplification reactions (set up according to therecommended protocol), primers are typically depletedbefore dNTPs. When DNA synthesis can no longertake place due to substrate depletion, subsequentrounds of DNA denaturation and annealing result in the6Technical Data Sheetseparation of complementary DNA strands, followed byimperfect annealing to non-complementary partners.This presumably results in the formation of so-called"daisy-chains" or "tangled knots", comprising largeassemblies of improperly annealed, partially doublestranded, heteroduplex DNA. These species migrateslower and are observed as secondary, highermolecular weight peaks during the electrophoreticanalysis of amplified libraries. However, they aretypically comprised of library molecules of the desiredlength, which are individualized during denaturationprior to cluster amplification or probe hybridization.Since these heteroduplexes contain significantportions of single-stranded DNA, over-amplificationleads to the under-quantification of library moleculeswith assays employing dsDNA-binding dyes. qPCRbased library quantifications methods, such as theKAPA Library Quantification assay, quantify DNA bydenaturation and amplification, thereby providing anaccurate measure of the amount of adapter-ligatedmolecules in a library, even if the library was overamplified.Please refer to the KAPA NGS Library PreparationTechnical Guide for a more detailed discussionof factors that can affect the efficiency of libraryamplification, and the impact of over-amplification onlibrary quantification. Excessive library amplification can result in otherunwanted artefacts such as amplification bias, PCRduplicates, chimeric library inserts, and nucleotidesubstitutions. The extent of library amplificationshould therefore be limited as much as possible, whileensuring that sufficient material is generated for QCand downstream processing (e.g. target enrichment orsequencing). If cycled to completion (not recommended) asingle 50 µl library amplification PCR, performedas described in Section 9, can produce8 – 10 µg (160 – 200 ng/µl) of amplified library.To minimize over-amplification and associatedundesired artefacts, the number of amplificationcycles should be optimized to produce an amplifiedlibrary with a concentration in the range of10 – 30 ng/µl (0.5 – 1.5 µg of PCR product per 50 µlreaction). Quantification of adapter-ligated libraries priorto library amplification can greatly facilitate theoptimization of library amplification parameters,particularly when a library construction workflow isfirst established. With the KAPA Library QuantificationKit, the amount of template DNA (adapter-ligatedmolecules) available for library amplification can bedetermined accurately. Using a simple calculation (forexponential amplification), the number of amplificationcycles needed to achieve a specific yield of amplifiedlibrary may be predicted theoretically (see Table 2 onthe next page).
KAPA LTP Library Preparation KitIllumina platformsLibrary amplification (continued) The actual optimal number of amplification cyclesmay be 1 - 3 cycles higher, particularly for librariesconstructed from FFPE DNA or other challengingsamples, or libraries with a broad fragment sizedistribution.Table 2. Theoretical number of cycles required to achieve0.5 – 1.5 µg (10 – 30 ng/µl) library in a standard 50 µl KAPA HiFiHotStart ReadyMix library amplification reaction, starting fromdifferent amounts of template DNA.Template DNANumber of cycles1 ng10 – 115 ng7–810 ng6–725 ng5–650 ng4–5100 ng3–4250 ng2–3500 ng0–2Evaluating the success of library construction Your specific library construction workflow should betailored and optimized to yield a sufficient amountof adapter-ligated molecules of the desired sizedistribution for the next step in the process (e.g. targetenrichment or sequencing), as well as for QC andarchiving purposes. The size distribution of the final or pre-capturelibrary should be confirmed with an electrophoreticmethod, whereas KAPA Library Quantification Kits forIllumina platforms are recommended for qPCR-basedquantification of libraries. These kits employ primersbased on the Illumina flow cell oligos, and can beused to quantify libraries:Technical Data Sheet Accurate quantification of DNA at this stage will allowyou to evaluate the efficiency of: the process from end repair to ligation, bydetermining the percentage of input DNA convertedto adapter-ligated molecules. library amplification with the selected number ofcycles, based on the actual amount of templateDNA used in the PCR. The availability of quantification data before and afterlibrary amplification allows the two major phases ofthe library construction process to be evaluated andoptimized independently to achieve the desired yieldof amplified library. If size selection is performed at any stage of theprocess, qPCR quantification before and after sizeselection may also be helpful to define the relativebenefit of size selection, and the loss of materialassociated with the
library amplification2, 3, 4. KAPA LTP Library Preparation Kits (KK8230 and KK8232) include KAPA HiFi HotStart ReadyMix (2X), a ready-to-use PCR mix comprising all the components for library amplification, except primers and template. Kits without an amplification module (KK8231 and KK8233) are available for PCR-free workflows.