Transcription
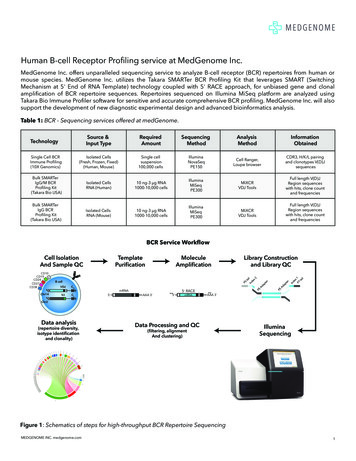
Human B-cell Receptor Profiling service at MedGenome Inc.MedGenome Inc. offers unparalleled sequencing service to analyze B-cell receptor (BCR) repertoires from human ormouse species. MedGenome Inc. utilizes the Takara SMARTer BCR Profiling Kit that leverages SMART (SwitchingMechanism at 5' End of RNA Template) technology coupled with 5' RACE approach, for unbiased gene and clonalamplification of BCR repertoire sequences. Repertoires sequenced on Illumina MiSeq platform are analyzed usingTakara Bio Immune Profiler software for sensitive and accurate comprehensive BCR profiling. MedGenome Inc. will alsosupport the development of new diagnostic experimental design and advanced bioinformatics analysis.Table 1: BCR - Sequencing services offered at medGenome.Source &Input formationObtainedSingle Cell BCRImmune Profiling(10X Genomics)Isolated Cells(Fresh, Frozen, Fixed)(Human, Mouse)Single cellsuspension100,000 cellsIlluminaNovaSeqPE150Cell Ranger,Loupe browserCDR3, H/K/L pairingand clonotypes V(D)JsequencesBulk SMARTerIgG/M BCRProfiling Kit(Takara Bio USA)Isolated CellsRNA (Human)10 ng-3 μg RNA1000-10,000 cellsIlluminaMiSeqPE300MiXCRVDJ ToolsFull length V(D)JRegion sequenceswith hits, clone countand frequenciesBulk SMARTerIgG BCRProfiling Kit(Takara Bio USA)Isolated CellsRNA (Mouse)10 ng-3 μg RNA1000-10,000 cellsIlluminaMiSeqPE300MiXCRVDJ ToolsFull length V(D)JRegion sequenceswith hits, clone countand frequenciesTechnologyBCR Service WorkflowTemplatePurificationCell IsolationAnd Sample brary Constructionand Library QCB cellVDJCHVJCLmRNA5'AAA 3'5'5' RACEmRNAAAA 3'Chr2Data analysis(repertoire diversity,isotype identificationand clonality)Data Processing and QC(filtering, alignmentAnd clustering)IlluminaSequencingFigure 1: Schematics of steps for high-throughput BCR Repertoire SequencingMEDGENOME INC. medgenome.com1
What is B-Cell Receptor (BCR) RepertoireAn adaptive immune system is fundamentally dependent upon the generation of diverse repertoires of B-lymphocytesantigen receptors (BCR). BCRs, a membrane bound cell surface receptors are assembled by genetic rearrangement andsomatic recombination of vast immunoglobulin gene segments and are constantly diversified to specifically bindexogenous antigens and endogenous host responses.V(D)J recombination is a process of tandem arrangement of variable (V), diversity (D) and joining (J) gene segments, thatoften results in nucleotide insertion and/or deletion between gene segments at the junction (Figure 2). Human BCRundergo recombination at variable region of three gene segments of the immunoglobulin heavy chain locus (V, D, J) andtwo gene segments of immunoglobulin light chain locus (V, J).The collection of B-cells receptors genetically rearranged for different antigen specificity are known as BCR repertoire.Vh1Vh2Vh3Vh40Dh1-23Jh1-6Ch1Ch2Ch3Stem cellsCh1Gene rearrangementCh2Ch3Naive B finity-matured B cellsineavCh1Somatic hypermutationand affinity maturationCh2Ch3Protein(immunoglobulin)Figure 2. Gene rearrangement in the B cell receptor Immunoglobulin Heavy ChainApplications of BCR Repertoire SequencingDiseaseDiagnosis osticMarkerDiscoveryB-cellRepertoireAnalysisFigure 3. BCR Repertoire ApplicationsMEDGENOME INC. medgenome.com2
Tracking known repertoire sequencesIncorporating repertoire sequencing into antibody discovery projects will enable the identification of antibodysequences that are either novel or previously identified. Such knowledge can aid in the development of new therapeuticstudies.Diagnostic Marker Discovery against Infectious DiseasesHigh throughput repertoire sequencing can provide broad information of disease-specific BCR clones and theirdynamic changes in clonality during an infectious state. Sequencing data from antigen-specific clones with stereotypedfeatures in the post-infection repertoires, can provide greater insights and contribute to diagnostic marker discoveryagainst infectious diseases such as H1N1, malaria and COVID-19.Disease Diagnosis and VaccinationRepertoire sequencing can provide insight into disease associated antibody repertoire information such as measure ofdiversity and rate of abundance of different antibody clone sequences. This repertoire sequence data can significantlyaid in understanding immunological mechanisms in vaccine development and improve our understanding in correlatingrepertoire sequence data to immunological assay functional measures.Major TechnologiesGene Amplification TechnologyTakara SMARTer Human BCR Profiling kit leverages SMARTtechnology (Switching Mechanism at 5’ End of RNA Template)and pairs NGS with a 5’ RACE approach (Figure 4A). cDNAsynthesis is dT-primed and full-length cDNA is achieved mplated nucleotides at 5’end of each mRNA template.The first strand cDNA is then subjected to two rounds mi-nested PCR. The nested PCR reduces variability and allowsfor priming from the constant region of heavy or light chains(Figure 4B). This method generates highly sensitive andreproducible B-cell repertoire profiling, and allow to capturecomplete V(D)J variable regions of BCR transcripts.Figure 4. Takara SMARTer Human BCR Profiling kit workflowMEDGENOME INC. medgenome.com3
Next Generation Sequencer (Illumina MiSeq)Nucleotide sequences of B-cell Receptor (BCR) genes require long readsequencing (400-600 bp) to read and assign the V, D, J and C regions ofthe BCR transcripts. For this purpose, BCR repertoire is sequenced usingIllumina MiSeq (Figure 5).Figure 5. Illumina MiSeqBCR Repertoire AnalysisMedGenome Inc. utilized Takara Bio Immune Profiler Software to analyze V, D, J and C regions of the BCR transcripts. Thesoftware incorporates two third-party software packages, MIGEC and MiXCR for accurate and reliable clonotype callingand quantification.A validation study at MedGenome Inc. was performed using total RNA from human spleen as well as B-cell expressingcell lines and PBMCs using the human Takara SMARTer BCR Profiling Kit. Sequencing data was analyzed using Takara BioImmune Profiler software following recommended guidelines. We provide a representative BCR repertoire report usinghuman spleen total RNA.erU741pper15001000700500400Size (bp)Size (bp)erppU0010wLo100015000700Size (bp)500Size 0002002000300010030004000504000500025Sample Intensity (Normalized FU)pp169UerwLo5000erDerCSample Intensity (Normalized FU)w0Size 0020020003000100300040005040005000LoSample Intensity (Normalized FU)U691pperw5000erBLoSample Intensity (Normalized FU)ASize (bp)Figure 6. TapeStation traces show representative profiles of B-cell receptor repertoire libraries generated from 10 ng oftotal RNA from human spleen to specifically amplify the IgG heavy chain (A), IgM heavy chain (B) Kappa (C) and Lambda(D) chains respectively. The libraries were generated using the Takara SMARTer Human BCR IgG IgM H/K/L Profiling Kitand following manufacturers’ instructions. Sequencing was performed using the Illumina MiSeqV3 600 cycle kit, andanalysis was performed using the immune profiler pipeline provided by Takara.MEDGENOME INC. medgenome.com4
Table 2. Mapping statistics shows specificity of amplification of heavy and light chains in BCR libraries of RNA fromhuman spleen total lHuman Spleen 10ng 3953(2.7%)0(0.0%)144175(100.0%)Human Spleen 10ng 93(2.6%)0(0.0%)53415(100.0%)Human Spleen 10ng 4257(2.7%)0(0.0%)156665(100.0%)Human Spleen 10ng )3783(3.6%)0(0.0%)106196(100.0%)Human Spleen 100ng .0%)59298(2.6%)0(0.0%)2297851(100.0%)Human Spleen 100ng %)14750(2.2%)0(0.0%)660269(100.0%)Human Spleen 100ng 0%)30286(2.6%)0(0.0%)1186967(100.0%)Human Spleen 100ng 0%)51057(3.2%)0(0.0%)1598277(100.0%)Table 3. Representative table of final clonotype counts from BCR libraries generated using 10 ng of total RNA humanspleen (Top 5 clonotypes) each amplifying the IgG heavy and light chains.Sample IDCloneIdCloneCountCloneFractionS3034351 IGLmig cdr3021S3034351 IGMmig cdr3011S3034352 IGGmig cdr30206S3034352 IGGmig cdr31S3034352 IGGmig cdr32S3034352 IGGmig cdr33S3034352 IGGmig cdr34MEDGENOME INC. medgenome.com113686845All VHitsWith ScoreIGLV2-14*00(2125),IGLV2-18*00(1874)IGHV3-7*00 (1462)0.009528655 IGHV3-48*00 (3179.4)0.0052268840.003145381IGHV3-7*00 (3466.7)IGHV3-74*00(3681)0.003145381 IGHV1-18*00(2714.6)0.002081502All DHitsWith 5),IGHD5-12*00(25)All JHitsWith ScoreAll CHitsWith ScoreAA Seq 0(80.2),CAGETYYYDHWIGHGP*00(78.2)5
Sample IDCloneIdCloneCountCloneFractionS3034352 IGGmig cdr35410.00189648S3034352 IGGmig cdr3S3034352 IGGmig cdr3S3034352 IGGmig cdr3S3034352 IGGmig cdr3S3034352 IGGmig cdr3S3034352 IGGmig cdr3678414039All VHitsWith ScoreIGHV4-59*00(3740.3),IGHV4-61*00(3534.5)All DHitsWith ScoreAll JHitsWith .001850224 IGHG2*00(85.6),IGHGP*00(83.3)IGHG1*00(73.5),9AA Seq l CHitsWith 99.5)IGHD3-22*00(74)IGHD1-1*00(41),11370.001711458 AKTARDWYD-IGHGP*00(89.2)EYWClonotype counts1500010000Antibody Chain5000IGGIGKIGLIGM010 ng100 ngHuman Spleen RNA InputFigure 7. Bar chart shows the total number of clonotypes identified in Human spleen RNA per Ig chain with differentconcentrations of RNA.MEDGENOME INC. medgenome.com6
Advanced DeliverablesIn addition to the standard deliverables using the MixCR pipeline and MedGenome’s pipeline for clonotypecomparisons, we also offer advanced bioinformatics and data visualization services using the V(D)J tools by HITIFDIVVTRRTWFDTWCATGEVTKHYYYGMDVW405060CDR3 length, HGSSGCLRWCARVLHYDVWSIYYYVLDVWOtherFigure 8. Sample BCR repertoire report with advanced deliverables including (A) Dendrograms and clustering diagramsshowing usage of the V(D)J genes across multiple samples, (B) Chord-diagram showing the usage and pairing of theBCR V(D)J genes (C) Spectratype plot of the CDR3 usage for the samples.MEDGENOME INC. medgenome.com7
ReferencesGeorgiou, G., Ippolito, G., Beausang, J. et al. The promise and challenge of high-throughput sequencing of theantibody repertoire. Nat Biotechnol 32, 158–168 (2014).Bolotin, D.A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015).Yaari, G. and Kleinstein, S.H. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med.7:121 (2015).Shugay M et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comp Biol 2015;11(11):e1004503-e1004503Profiling mouse B-cell receptors with SMART technology, Takara Bio. Retrieved -(bulk).MEDGENOME INC. medgenome.com8
MMLV-derived SMARTScribe Reverse Transcriptase (RT) enzyme, that adds SMART UMI oligo’s annealed to non-templated nucleotides at 5’end of each mRNA template. The first strand cDNA is then subjected to two rounds of gene-specific PCR amplification are performed using semi-