Transcription
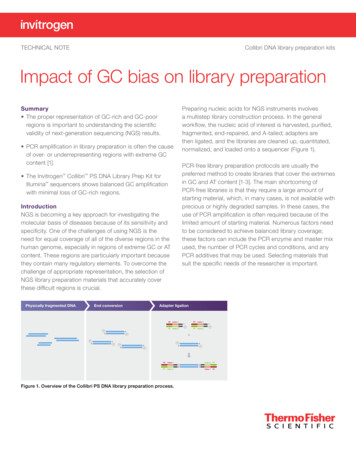
TECHNICAL NOTECollibri DNA library preparation kitsImpact of GC bias on library preparationSummary The proper representation of GC-rich and GC-poorregions is important to understanding the scientificvalidity of next-generation sequencing (NGS) results. PCR amplification in library preparation is often the causeof over- or underrepresenting regions with extreme GCcontent [1]. The Invitrogen Collibri PS DNA Library Prep Kit forIllumina sequencers shows balanced GC amplificationwith minimal loss of GC-rich regions. IntroductionNGS is becoming a key approach for investigating themolecular basis of diseases because of its sensitivity andspecificity. One of the challenges of using NGS is theneed for equal coverage of all of the diverse regions in thehuman genome, especially in regions of extreme GC or ATcontent. These regions are particularly important becausethey contain many regulatory elements. To overcome thechallenge of appropriate representation, the selection ofNGS library preparation materials that accurately coverthese difficult regions is crucial.Figure 1. Overview of the Collibri PS DNA library preparation process.Preparing nucleic acids for NGS instruments involvesa multistep library construction process. In the generalworkflow, the nucleic acid of interest is harvested, purified,fragmented, end-repaired, and A-tailed; adapters arethen ligated, and the libraries are cleaned up, quantitated,normalized, and loaded onto a sequencer (Figure 1).PCR-free library preparation protocols are usually thepreferred method to create libraries that cover the extremesin GC and AT content [1-3]. The main shortcoming ofPCR-free libraries is that they require a large amount ofstarting material, which, in many cases, is not available withprecious or highly degraded samples. In these cases, theuse of PCR amplification is often required because of thelimited amount of starting material. Numerous factors needto be considered to achieve balanced library coverage;these factors can include the PCR enzyme and master mixused, the number of PCR cycles and conditions, and anyPCR additives that may be used. Selecting materials thatsuit the specific needs of the researcher is important.
In general, if samples are identified as being challenging,the easiest factors to modulate are the number of PCRcycles, the cycling conditions, and the addition of PCRenhancers. An increase in the number of PCR cycles ina protocol usually increases any bias caused by the PCRenzyme and master mix used; the recommended approachis therefore to minimize the number of cycles while ensuringthat sufficient product is loaded onto the sequencer. Fourto eight PCR cycles is typical with this approach.A greater number of cycles and a higher annealingtemperature tend to increase the specificity of theamplification protocol; however, the increase in specificityis achieved at the expense of losing regions with extremeAT content. Thus, the use of mid-range annealingand extension temperatures, such as 60 C and 72 C,respectively, is important. The PCR enzyme and mastermix used may exert the greatest influence on the GCcoverage of the library preparation [1-3].MethodsLibraries were prepared using four different librarypreparation kits—the Collibri PS DNA Library Prep Kit forIllumina Systems, and older library prep kits including theKAPA HyperPrep Kit, NEBNext Ultra II DNA Library PrepKit, and TruSeq DNA Nano Library Preparation Kit. Allsteps were performed according to the manufacturers’protocols. The NGS libraries were prepared usinggenomic DNA from the Coriell Institute for MedicalResearch (accession number NA12878), and with theHorizon Quantitative Multiplex Formalin Compromised(Moderate) Reference Standard (Cat. No. HD799). Forcomparison, 100 ng DNA samples of both types wereused. Samples were sequenced on the NovaSeq 6000Sequencing System with an S4 flow cell. GC bias wascalculated using Picard tools v2.7.1. Formalin-compromisedDNA was converted into sequencing libraries using themanufacturers’ recommended protocols and sequencedat 2 x 150 bp using unique dual 8-base indexes forsample identification.
ResultsThe sequencing run resulted in 85% Q30 bases forboth read 1 and read 2. The samples were normalizedto 25x coverage and analyzed. The resulting librarieswere analyzed for specific GC content as described[2]. In general, the Collibri and KAPA kits gave the mostconsistent coverage of extreme GC regions (Figure 2).The Collibri kit also showed the highest mean coveragefor “bad promoters”and areasof the genome withrage improvesgenomeassemblieswithout the need to increase sequencing depth;greater than 75% GC content. Overall, the Collibriary Prep Kits provide the most consistently even coverage among all input levelslibrary preparation kit resulted in consistent, precisegenomic coverage.mic Interpretations From Genomic InterpretationAGC coverage of 100 ng of genomic DNAedictable GC coverage improves genome assemblies without the need to increase sequencing depth;Collibri DNA Library Prep Kits provide the most consistently even coverage among all input levelsFFPE PerformanceA Library Prep Kitrep KitII Library Prep KitCollibri PCR-Free PS DNA Library Prep KitCoverage of challenging regions doesnot detract from exon coverageKAPA Hyper Prep KitCollibri PS DNA Library Prep KitKAPA Hyper Prep KitII LibraryPrep KitCollibri PSNEBNextDNA UltraLibraryPrepKitTruSeq Nano DNA Library Prep KitKAPA HyperPrepKitCollibriPS DNALibrary Prep KitNEBNext Ultra II Library Prep KitCollibri PCR-Free PS DNA Library Prep KitTruSeq DNA PCR-Free Library Prep KitTruSeq Nano DNALibrary Prep KitKAPA HyperNEBNextPrep KitUltra IIKAPA Hyper Prep KitPrep KitTruSeq DNALibraryPCR-FreeLibrary Prep Kitequenced on an Illumina NovaSeq 6000 System and reads normalized to 186M reads. GC bias was calculated using Picard tools v2.7.1.BTruSeqNanoregion:DNA Library Prep KitChallenging“bad promoters”Challenging region:75% GCExon coverage6000 System and reads normalizedto 186M reads. GC bias was calculated3.0 using Picard tools v2.7.1.3.01.5FFPE Performance1.0Normalized coverage2.0Normalized coverageNormalized Coverage of challenging regionsdoes not detract from exon coverage0.00.0Collibri PS DNATruSeq Nano DNAKAPA Hyper Prep KitNEBNext Ultra IILibraryPrepKitconverted into sequencingLibraryPreptheKitmanufacturer’s recommended protocols and sequenced on an Illumina NovaSeqLibrary 6000PrepSystemKit with a 2 x 150 bp read length.100 ng of Horizon moderate damageFFPE DNAwerelibraries usingResulting PF reads were normalized to 186M reads.Challengingregion:coverage of challenging Challengingregion:genome. (A) Normalized coverage of the percent of mappedFigure 2. Graphsshowing normalizedregions in the humanExon coverage12promoters”75%challengingGCGC content of 100 “badng of CoriellNA12878 DNA. (B) Coverage of promoters withGC content and 75% GC content in Horizon FFPE DNA ishigher and more even with the Collibri PS DNA Library Prep Kit. “Bad promoters” include 1,000 GC-rich human promoters that are exceptionally resistant3.03.0to sequencing [2]. 3.01.02.01.51.0ormalized coverage1.5ormalized coverageormalized coverage2.02.52.52.52.01.51.0
Conclusions Balanced coverage of the diverse regions in the humangenome is important for the investigation of the molecularbasis of diseases. PCR amplification of libraries prepared with older libraryprep technology can lead to coverage bias in regionswith extreme GC and AT content. The Collibri PS DNA Library Prep Kit enables consistentgenomic coverage.Ordering informationProductQuantityCat. No.with CD Indexes24 prepsA38605024with CD Indexes96 prepsA38607096with UD Indexes, Set A (1-24)24 prepsA38606024with UD Indexes, Set B (25-48)24 prepsA43605024with UD Indexes, Set C (49-72)24 prepsA43606024with UD Indexes, Set D (73-96)24 prepsA43607024with CD Indexes24 prepsA38545024with CD Indexes96 prepsA38603096with UD Indexes, Set A (1-24)24 prepsA38602024with UD Indexes, Set B (25-48)24 prepsA43602024with UD Indexes, Set C (49-72)24 prepsA43603024with UD Indexes, Set D (73-96)24 prepsA43604024with CD Indexes24 prepsA38612024with CD Indexes96 prepsA38614096with UD Indexes, Set A (1-24)24 prepsA38613024with UD Indexes, Set B (25-48)24 prepsA43611024with UD Indexes, Set C (49-72)24 prepsA43612024with UD Indexes, Set D (73-96)24 prepsA43613024with UD Indexes, Set A-D (1-96)96 prepsA38614196with CD Indexes24 prepsA38608024with CD Indexes96 prepsA38610096with UD Indexes, Set A (1-24)24 prepsA38609024with UD Indexes, Set B (25-48)24 prepsA43608024with UD Indexes, Set C (49-72)24 prepsA43609024with UD Indexes, Set D (73-96)24 prepsA43610024with UD Indexes, Set A-D (1-96)96 prepsA38615196DNA-Seq kits for Illumina systemsCollibri ES DNA Library Prep KitsCollibri PCR-Free ES DNA Library Prep KitsCollibri PS DNA Library Prep KitsCollibri PCR-Free PS DNA Library Prep KitsCD combinatorial dual, UD unique dual
Ordering information (continued)ProductQuantityCat. No.24 prepsA3899402496 prepsA3899409624 prepsA39003024RNA-Seq kits for Illumina systemsCollibri Stranded RNA Library Prep Kit for Illumina SystemsCollibri Stranded RNA Library Prep Kit for Illumina Systems with H/M/R rRNA Depletion Kit96 prepsA39003096ERCC RNA Spike-In Mix1 kit4456740ERCC ExFold RNA Spike-In Mixes1 kit4456739100 rxnsA38524100Library quantificationCollibri Library Quantification Kit500 rxnsA38524500Qubit 4 Fluorometer, with WiFi1 fluorometerQ33238Qubit 4 NGS Starter Kit, with WiFi1 kitQ3324050 rxnsA38539050250 rxnsA3853925050 rxnsA38540050250 rxnsA38540250Library amplificationCollibri Library Amplification Master MixCollibri Library Amplification Master Mix with Primer MixH/M/R human/mouse/ratReferences1. Aird D, Ross MG, Chen WS et al. (2011) Analyzing and minimizing PCR amplificationbias in Illumina sequencing libraries. Genome Biol 12:R18.2. Ross MG, Russ C, Costello M et al. (2013) Characterizing and measuring bias insequencing data. Genome Biol 14:R51.3. Chen Y-C, Liu T, Yu C-H et al. (2013) Effects of GC bias in next-generation sequencingdata on de novo genome assembly. PLoS One 8:e62856.Find out more at thermofisher.com/collibriFor Research Use Only. Not for use in diagnostic procedures. 2019 Thermo Fisher Scientific Inc. All rights reserved. Alltrademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. TruSeq and NovaSeq aretrademarks of Illumina Inc. KAPA is a trademark of Roche. NEBNext is a trademark of New England BioLabs Inc. Horizon is a trademarkof Horizon Discovery Group PLC. The following DNA samples were obtained from the NIGMS Human Genetic Cell Repository at theCoriell Institute for Medical Research: NA12878. COL23554 0919
Library Prep Kit KAPA Hyper Prep Kit NEBNext Ultra II Library Prep Kit Figure 2. Graphs showing normalized coverage of challenging regions in the human genome. (A) Normalized coverage of the percent of mapped GC content of 100 ng of Coriell NA12878 DNA. (B) Coverage of promoters with challenging GC content and 75% GC content in Horizon FFPE DNA is