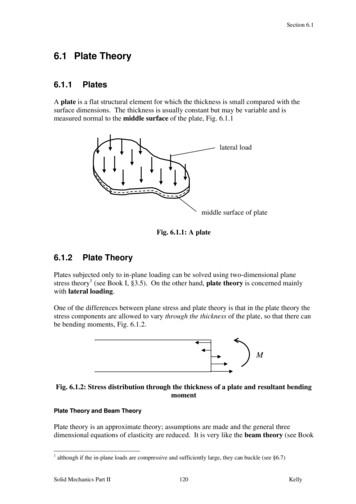
Transcription
Theory in Practice: Modelingin NeuroimagingHow to model “big” MRI datasets
Outline of talk Theory recap: modelling approaches can be reduced to two types:predictive and descriptive “Big data” complicates our ability to apply both approaches Marginal Modelling is a good approach good for descriptive modelling Functional Random Forests is a good approach for predictivemodelling Other approaches can also handle big data, but are beyond the scopeof this workshop
Before even considering models, we need toknow what question to ask How and where may cortical thickness be associated with workingmemory performance?
Before even considering models, we need toknow what question to ask How and where may cortical thickness be associated with workingmemory performance? Can measures of functional brain organization predict an individual’sworking memory ability?
Each question requires a different modellingapproach How and where may cortical thickness be associated with workingmemory performance? Descriptive modelling Can measures of functional brain organization predict an individual’sworking memory ability? Predictive modelling
Descriptive models measure what one hascollected predictive models measure what one ytics-vs-descriptive-analytics/
Descriptive models explore data, predictivemodels confirm properties of s-descriptive-analytics/
Descriptive models provide insight, predictivemodels apply s-vs-descriptive-analytics/
Descriptive models are limited to in-sample data,predictive models require out-of-sample s-descriptive-analytics/
Descriptive models are assessed via theory andinference, predictive models are assessed byindependent cs-vs-descriptive-analytics/
Outline of talk Theory recap: modelling approaches can be reduced to two types:predictive and descriptive “Big data” complicates our ability to apply both approaches Marginal Modelling is a good approach for descriptive modelling Functional Random Forests is a good approach for predictivemodelling Other approaches can also handle big data, but are beyond the scopeof this workshop
First, all health-focused imaging studiesshould probably be big 31141-8.pdf
Our ABCD pipeline generates anywhere from10 to 90 thousand )31141-8.pdf
Our ABCD pipeline generates anywhere from 10 to90 thousand tests (some special cases are 273(17)31141-8.pdf
We’ve collected about 10,000 )31141-8.pdf
ABCD needed a lot of coordination and dataaggregation to collect over 10,000 participantsAuchter et al, 2018, https://doi.org/10.1016/j.dcn.2018.04.003
Descriptive models must take into accountthis nested structure Complex models may be slow to calculate when analyzing 4500participants Permutation tests may take days or even weeks Permutation tests lack exchangeability for complex questions
Permutation testing can reveal whetherdifferences in community structure aresignificantly differentdepressionHirschhorn,2005, https://doi.org/10.1038/nrg1521
Permute group assignment and calculatestatisticdepressionno depression‘depression’‘no depression’Hirschhorn,2005, https://doi.org/10.1038/nrg1521
Do so for multiple permutations and construct adistribution of the statistic for permuted groupsdepressionno depression‘depression’‘no depression’Hirschhorn,2005, https://doi.org/10.1038/nrg1521
FrequencyP value is determined by the proportional rankof the observed statistic compared to thepermuted distribution
At a Z 2.3, falsepositive rates are highwhen not usingpermutation testing
At a Z 3.1, falsepositive rates aregenerally better andin-line with the trueFP rate
This all works because each individual isindependently acquired from one another – thedata are exchangeable
Independence gets more complicated when youhave more complicated designs – but even herewe can exchange every individualDrug useCannabisAlcoholNicotine StimulantAnderson and Braak, 2003, JSCS; 10.1080 0094965021000015558
However, if a second factor is nested, ourpermutations are limited to the nested pairs,restricting our permutationsDrug useCannabis Alcohol Nicotine StimulantFamily nested by drug useAnderson and Braak, 2003, JSCS; 10.1080 0094965021000015558
More complex designs have even morerestrictions, relative to the total number ofpermutationsDrug useHometownCannabis Alcohol Nicotine StimulantAnderson and Braak, 2003, JSCS; 10.1080 0094965021000015558
In turn, restricted permutations have reducedpower when controlling for the false positive rateAnderson and Braak, 2003, JSCS; 10.1080 0094965021000015558
Predictive models must also take into accountnested /PMC5736019/
Scanner effects can be common, independentof siteGareth Harman, 4/11/19 – combat Cortical Thickness
ComBat has also been used to correct forABCD data, which can be predicted by siteSite classificationaccuracyNielson, 2018, biorxiv; http://dx.doi.org/10.1101/309260
Cross-validation strategies can mitigateknown but not unknown effects Stratified validation is possible via independent stratified groups Leave-one-site-out validation can help catch site effects But what about effects of scanner upgrades, software maintenance,or even changes in personnel?
Outline of talk Theory recap: modelling approaches can be reduced to two types:predictive and descriptive “Big data” complicates our ability to apply both approaches Marginal Modelling is a good approach for descriptive modelling Functional Random Forests is a good approach for predictivemodelling Other approaches can also handle big data, but are beyond the scopeof this workshop
The marginal model may be a more feasiblesolution for modeling ABCD populations Strengths: Marginal model makes few assumptions with respect to the data Nested-designs can be modeled or unmodeled, and left to the error term (hopefully) Individual cases can be incomplete or missing for a marginal model Longitudinal designs are feasible within the marginal model framework Marginal model has a closed-form solution to the equation via a SandwichEstimator (SwE) It’s fast, and can be feasibly run with limited resources on lots of data Use of a wild bootstrap (WB) provides an NHST framework for complexquestions
Critical limitations The marginal model cannot be used to draw inferences aboutindividuals within a population It is an exploratory approach, which can be verified using subsequentconfirmatory approaches DEAP can help conform such analyses to best standards and practices throughpre-registered reports, reproducibility, and independent validation
Bryan Gillaume’s and Tom Nichols implemented anapproach that uses a sandwich estimator to solvea marginal modelEstimate FEcovariance(SwE)ComputemodelY/X BetaPerformsmallsample adj.PerformWald TestStatistical T mapfor ect/groupscovariance(residuals)
Marginal models are effectively linear, so we firstestimate the parameters for our design matrix bydividing the imaging measure (Y) by the design (X)ComputemodelY/X BetaDesignmatrixImagingVolume(s)
For our software, the design matrix is justyour non-imaging dataComputemodelY/X BetaDesignmatrixImagingVolume(s)
So for example, with the ABCD data we caninput measures and test a modelMarginal model: y RTComputemodelY/X BetaDesignmatrixImagingVolume(s)
A sandwich estimator is used to estimatecovariance and determine the fixed effectsparametersEstimate FEcovariance(SwE)ComputemodelY/X BetaDesignmatrixImagingVolume(s)
To handle nested structure, group covariance canbe calculated separately (CRITICAL FOR ABCD)Estimate FEcovariance(SwE)ComputemodelY/X roupscovariance(residuals)
For ABCD, it is good to control for site andgenderEstimate FEcovariance(SwE)5ComputemodelY/X ctgender/groups142covariance(residuals)2
If needed we can perform a small sample sizeadjustment – this may be important if we usedfamily as a nesting variableEstimate FEcovariance(SwE)ComputemodelY/X roupscovariance(residuals)Performsmallsample adj.
Finally, a Wald test extracts a t-map forstatistical inferenceEstimate FEcovariance(SwE)ComputemodelY/X BetaPerformsmallsample adj.PerformWald TestStatistical T mapfor ect/groupscovariance(residuals)
The statistical map looks like thisEstimate FEcovariance(SwE)ComputemodelY/X BetaPerformsmallsample adj.PerformWald TestStatistical T mapfor ect/groupscovariance(residuals)
Use of a wild bootstrap enables inference similarto a permutation test – so we can control for theFWEREstimate (residuals)Performsmallsample adj.ComputemodelY/X BetaStatistical T mapfor inferenceDesignmatrixImagingVolume(s)PerformWald TestWB mapsWildbootstrapClusterdetection/TFCEInference map
Such a test allows us to detect significantclustersEstimate (residuals)Performsmallsample adj.ComputemodelY/X BetaStatistical T mapfor inferenceDesignmatrixImagingVolume(s)PerformWald TestWB mapsWildbootstrapClusterdetection/TFCEInference map
Wild bootstrap WB value fitted value residual value*sample value Sample with replacement can be from simple or complexdistributions: Radenbacher (-1, 1) would mean we either: WB value fitted value – residual value WB value fitted value residual value However, LOTS of possible distributions, so choice of distribution isimportant.
We have begun to implement a standaloneMarginalModelCifti package in RAlpha version will be released at -- http://github.com/dcan-labs/MarginalModelCifti
The main wrapper for MarginalModelCifti takes inimaging volumes and prepares them for analysisPrepCIFTI/Surf/VolImagingVolume(s)
ComputeMM is applied to the prepared data; userspecifies the model using Wilkinson notation andwraps the SwE and Wald Test using GeepackY group MMStatistical T mapfor inference
ComputeMM WB generates the WB mapsused to draw inferences about the T istical T mapfor inferenceComputeMM WBNull Distribution
In turn a family of functions are used toparallellize ComputeMM stical T mapfor inferenceComputeMM WBNull DistributionApplyWB to siudalsGetVolAreas
Cluster detection is performed within the mainwrapper, using information from both processesPrepCIFTI/Surf/VolComputeMMComputeMM WBStatistical T mapfor inferenceNull DistributionClusterdetection/TFCEInference mapImagingVolume(s)
The MarginalModelCifti packagecomprises multiple functions that canbe accessed by anyone
Functions are documented in accordancewith CRAN guidelines
Here are all the parameters for ConstructMarginalModel()
To make things easier – we’ve made a jupyternotebook that can be used as a reference
Outline of talk Theory recap: modelling approaches can be reduced to two types:predictive and descriptive “Big data” complicates our ability to apply both approaches Marginal Modelling is a good approach for descriptive modelling Functional Random Forests is a good approach for predictivemodelling Other approaches can also handle big data, but are beyond the scopeof this workshop
Nested structures -- people belong tomultiple subtypesDialect preferences: soda, coke or pop?SODACOKEPOPFeczko, Miranda-Dominguez, Marr, Graham, Nigg, Fair, TICS, 2019, DOI: https://doi.org/10.1016/j.tics.2019.03.009
Nested structures -- people belong tomultiple subtypesU.S. 2016 presidential election voting preferencesDialect preferences: soda, coke or pop?SODACOKEPOPDEMGOPFeczko, Miranda-Dominguez, Marr, Graham, Nigg, Fair, TICS, 2019, DOI: https://doi.org/10.1016/j.tics.2019.03.009
Nested structures -- people belong tomultiple subtypesU.S. 2016 presidential election voting preferencesDialect preferences: soda, coke or pop?SODACOKEPOPDEMStroke mortality for Adults 35 per 100,000GOPRATEFeczko, Miranda-Dominguez, Marr, Graham, Nigg, Fair, TICS, 2019, DOI: https://doi.org/10.1016/j.tics.2019.03.009
But what about effects of scanner upgrades,software maintenance, or even changes inpersonnel?
If we want to control for unknown structure, weneed to identify subtypes tied to an outcome Supervised approaches can confirm known subtypes but not discoverunknown subtypes tied to an outcome
If we want to control for unknown structure, weneed to identify subtypes tied to an outcome Supervised approaches can confirm known subtypes but not discoverunknown subtypes tied to an outcome Unsupervised approaches can discover unknown subtypes, but nottied to any outcome
How does the Functional Random Forestwork?Supervised component
Ask a question: can we predict depressiondiagnosis?Supervised componentUnsupervised component
We start with an input datasetInput datasetSupervised componentUnsupervised component
We start with an input datasetInput datasetSupervised componentUnsupervised component
This dataset can be a functional connectivitymatrixInput datasetSupervised componentUnsupervised component
This dataset can be a functional connectivitymatrix – which gets reduced to either graphmetrics or principal componentsInput datasetSupervised componentUnsupervised component
Input data are modeled via a random forestvia validation/testingInput datasetSupervised componentUnsupervised componentRandom ForestCreates decision trees
Model is supervised because it attempts topredict the outcome of interestInput datasetSupervised componentUnsupervised componentRandom ForestCreates decision trees
If the random forest performs well onindependent test data, a similarity matrix isproduced from the RFsInput datasetSupervised componentRandom ForestCreates decision treesSimilarity matrixUnsupervised component
Subgroups are identified from this matrix viaInfomapInput datasetSupervised componentRandom ForestCreates decision treesSimilarity matrixUnsupervised componentInfomapIdentifies communities
Subtypes arise from the model that are tiedto the outcomeInput datasetSupervised componentRandom ForestCreates decision treesSimilarity matrixUnsupervised componentInfomapIdentifies communitiesSubpopulations
The FRF can be used to identify trajectories inlongitudinal dataLongitudinal datasetFunctional Data AnalysisGenerates individualtrajectoriesf(t) a1ø1(t) . akøk(t)
Combining the set of functions estimates asmooth trajectory for an individual’s symptomsLongitudinal datasetFunctional Data AnalysisGenerates individualtrajectoriesf(t) a1ø1(t) . akøk(t)
Combining the set of functions estimates asmooth trajectory for an individual’s symptomsLongitudinal datasetFunctional Data AnalysisGenerates individualtrajectoriesf(t) a1ø1(t) . akøk(t)
We can use an unsupervised approach toidentify trajectoriesUnsupervisedLongitudinal datasetCorrelation MatrixCompares trajectoriesFunctional Data AnalysisGenerates Correlation-basedsubpopulationsf(t) a1ø1(t) . akøk(t)
Or use a “hybrid” approach that identifiestrajectory subtypes tied to an outcome of interestUnsupervisedLongitudinal datasetHybridParametersCorrelation MatrixCompares trajectoriesRandom ForestCreates decision treesFunctional Data AnalysisGenerates Correlation-basedsubpopulationsf(t) a1ø1(t) . akøk(t)Similarity pulations
A manual for using the FRF exists omforest/)
A new release is available at:
A manual for using the FRF exists omforest/)
Outline of talk Theory recap: modelling approaches can be reduced to two types:predictive and descriptive “Big data” complicates our ability to apply both approaches Marginal Modelling is a good approach for descriptive modelling Functional Random Forests is a good approach for predictivemodelling Other approaches can also handle big data, but are beyond thescope of this workshop
New approaches within statistics and machinelearning can also accommodate problems with bigdata Many of these approaches have been developed in genomics comBat is a Bayesian approach to handle known site effects in data Surrogate Variable Analaysis Such approaches need to be examined in the context of neuroimagingdata to evaluate where each is most useful Knowing how to use these tools requires considerable skill in datascience, which has been relatively untaught in mental health fields Hopefully, the workshop tomorrow should get you excited aboutapplying these new tools and on your path towards doing “big data”science right.
AcknowledgmentsFair Lab-Damien Fair-Oscar Miranda-Dominguez-Alice GrahamAlpha Testers-Bene Ramirez-Jennifer Zhu-Robert Hermosillo-Mollie Marr-Oliva Doyle-Michaela Cordova-AJ MitchellComputing Team-Darrick Sturgeon-Eric Earl-Anders Perrone-Emma Schifsky-Anthony Galassi-Kathy Snider-David Ball-Lucille Moore
Acknowledgments The mentors The databasors The developers Damien Fair Lourdes Irwin Eric Earl The assessors: Joel Nigg Darrick Sturgeon Anders Perrone Beth Langhorst Eric Fombonne Rachel Klein Darrick Sturgeon Michaela Cordova Shannon McWeeney Bene Ramirez Brian Mills Olivia Doyle Other Labs: Nigg Lab The students: McWeeney Lab Iliana Javier Nadir Balba The collaborators: The docs: Sarah Karalunas Alice Graham Alison Hill Oscar Miranda-Dominguez Jan Van Santen Binyam Nardos Everyone I forgot, which is many
Questions?
High dimensionality is bad for predictivemodellingFeczko, Miranda-Dominguez, Marr, Graham, Nigg, Fair, TICS, 2019, DOI: https://doi.org/10.1016/j.tics.2019.03.009
Predictive models must also take into accountnested /PMC3880143/
Theory in Practice: Modeling in Neuroimaging . Descriptive models are assessed via theory and inference, predictive models are assessed by independent testing . Statistical T map for inference Estimate FE covarian