Transcription
Introduction to MRI dataprocessing with FSLAnna Blazejewska
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduFSL FMRIB Software Library FMRIB Functional Magnetic Resonance Imaging of the Brain @ Oxfordsince 2000, last stable FSL 5.0, free!for structural MRI, functional MRI (task, resting), diffusion MRIdata processing & analysis written in C & TCL for Linux (virtual box on Windows) & Mac OSGUI but also command line!! shell script pipelinesinstallation: fslinstaller.pysupport: wiki, FAQ, forum yearly FSL courses: theory & hands on, 5 days, slides on-linehttp://fsl.fmrib.ox.ac.uk/fslcourse/
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edustructural MRI registration: linear (FLIRT) & non-linear (FNIRT)brain segmentation/extraction (BET)tissue type segmentation (FAST)subcortical structures segmentation (FIRST)voxelwise GM density analysis (FSLVBM)atrophy estimation (SIENA)functional MRI motion correction (MCFLIRT)EPI distortion correction (FUGUE, PRELUDE)model-based analysis (FEAT)model free ICA-based analysis (MELODIC)Bayesian analysis of perfusion, ASL data (FABBER, VERBENA, BASIL)diffusion MRIFSL overview distortion correction (TOPUP)eddy current correction (EDDY)diffusion toolbox (FDT)tract-based spatial statistics (TBSS) general: fslmaths, fslchfiletype, fslroi, smoothing, stats, fslview and more
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edustructural MRI registration: linear (FLIRT) & non-linear (FNIRT)brain segmentation/extraction (BET)tissue type segmentation (FAST)subcortical structures segmentation (FIRST)voxelwise GM density analysis (FSLVBM)atrophy estimation (SIENA)functional MRI motion correction (MCFLIRT)EPI distortion correction (FUGUE, PRELUDE)model-based analysis (FEAT)model free ICA-based analysis (MELODIC)Bayesian analysis of perfusion, ASL data (FABBER, VERBENA, BASIL)diffusion MRIFSL overview distortion correction (TOPUP)eddy current correction (EDDY)diffusion toolbox (FDT)tract-based spatial statistics (TBSS) general: fslmaths, fslchfiletype, fslroi, smoothing, stats, fslview and more 1.2.3.4.segmentationregistrationdistortion correctionshell scripts! fsl
1. segmentation
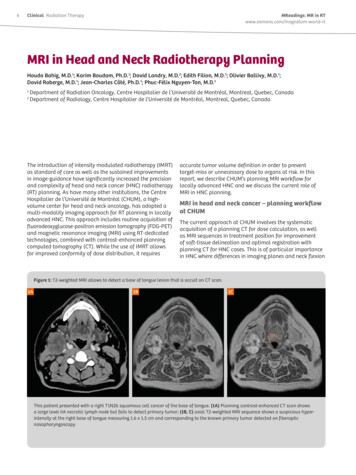
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu1. segmentationBET: brain extraction skull stripping goal: automatic segmentation of brain & non-brain tissue(skin, skull, eyeballs, etc.) preparation for: registration/motion correction tissue segmentation masking out non-brain problems of manual segmentation: time, training, reproducibility benefits of BET 5-20 sec, high reproducibility different contrasts T1-/T2-/T2*-w data, robust to bias field (uses local intensity changes) can estimate inner & outer skull & outer scalp surfaceSmith, SM, Fast robust automated brain extraction, HBM 17(3), 2002.Jenkinson, M et al., BET2: MR-based estimation of brain, skull and scalp surfaces, OHBM, 2005.brainnon-brain
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduBET: how does it work?1. segmentationsurface meshof connected triangles(no folds, not cortex)histogram-basedthreshold t estimationsubdividing each triangleexpanding the surfacevertex locations updatedbased on local intensitiest-based binarizationcenter of gravity (COG)not self-intersectingsurface smoothness conditionspherical surfaceinitializationSmith, SM, Fast robust automated brain extraction, HBM 17(3), 2002.
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduBET: how to use it? bet input output [parameters]-f f -g g f brain outline estimate, [0,1], 0.5 g brain outline bottom, top, [-1,1], 07T Philips MPRAGE 1.0 mm3default: f 0.5 g 0 modified: f 0.3 g 0.31. segmentation
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduBET: how to use it? bet input output [parameters]-c x y z -r r initial center of gravity [voxels]head radius [mm], initial r/2-R-Z-Frobust brain center estimationtemporary add slices for small z-FOVapply to 4D fMRI data (-f 0.3 dilation)-m-s-o-n-Asave binary brain masksave skull imagesave brain surface outlineskip segmented brain imagegenerate skull & scalp surfaces (BETSURF)1. segmentation
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduFAST: tissue segmentationFMRIB’s Automated Segmentation Tool1. segmentationhttp://fsl.fmrib.ox.ac.uk/fslcourse goal: automatic segmentation of WM, GM & CSF input: BET processed images (extracted brain) single image: T1, T2, PD multichannel images pre-aligned (with FLIRT) output: binary tissue masks or probability maps1.2.3.4.histogram-based: uses Gaussian mixture modelestimates & removes bias fieldconsiders voxel’s neighborhood robust to noiseuse of prior tissue probability mapsGMZhang, Y et al., Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm, IEEE Trans Med Imag, 20(1), 2001.CSFWM
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu1. segmentationFAST: histogram as a mixture of Gaussians data intensity histogrammodel mixture of Gaussiansseparation of the peaks segmentationpeaks overlap segmentation more difficultfor each voxel calculates: P(CSF), P(GM), P(WM)GM WMCSFWMGMCSFP(CSF) 0P(GM) 400P(WM) fmrib.ox.ac.uk/fslcourseGM/WM border
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu1. segmentationFAST: hard segmentation vs probability mapsoriginalimage3 classsegmentation7T Siemens MEMPRAGE 0.75 mm3CSFprobability mapsGMWM
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu1. segmentationFAST: bias field correction low resolution, head motion, noise, blurring & bias field histogram peaks overlap RF inhomogeneity spatial intensity variations bias fieldhttp://fsl.fmrib.ox.ac.uk/fslcourseoriginal imageestimated bias fieldcorrected image
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu1. segmentationFAST: neighborhood local neighborhood information robust to noise P(class) P(intensity) P(neighborhood) controls contribution of neighbors vs intensity, can be set by user 0http://fsl.fmrib.ox.ac.uk/fslcourse 0.1 0.3 0.5
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduFAST: how to use it? fast [options] input-H-t-S , 0.3input data type: 1 T1, 2 T2, 3 PDnumber of input data channels, 1-b-B-N-I-lsave estimated bias fieldsave bias-corrected input imageno bias field correctionnumber of iteration for bias field removal, 4bias field smoothing FWHM, 20mm-n-g-nopvenumber of tissue type classes, 3save a binary mask for each classskip probability maps1. segmentation
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu1. segmentationFAST: use of priors-a t.mat-P-A p1 p2 p3-s fileuse prior probability maps for initialization(requires transformation to standard space)use prior probability maps at all stages (requires –a/-A)alternative prior images for tissue classesinitial tissue-type meansT1-w meanGMWMCSFhttp://fsl.fmrib.ox.ac.uk/fslcourse
2. registration
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu2. registrationregistration transformation of 3D volumes into the same space (coordinate system)1. transformation 2. cost function 3. interpolation intra-subject: different modalities, contrasts, frames etc. inter-subject: different subjects to common space (population analysis etc.)MNI152 152 structural images, averagedafter high-dimensional nonlinear registrationMNI152lin T1 1mm @ FSLDIR/data/standardsJenkinson M & Smith. A global optimisation method for robust affine registration of brain images. Medical Image Analysis, 5(2):143-156, 2001.M. Jenkinson, P.R. Bannister, J.M. Brady, and S.M. Smith. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage, 17(2):825-841, 2002.
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu2. registrationFLIRT: linear transformationFMRIB’s Linear Registration Tool intra-subject, same/different modality, DOF degrees of freedomhttp://fsl.fmrib.ox.ac.uk/fslcourserigid body 6DOF 3 rotations 3 translationsaffine 12 DOF 6DOF 3 scalings 3 skews/shears represented by a transformation matrix𝑎11 𝑎12 𝑎13 𝑎14𝑎21 𝑎22 𝑎23 𝑎24𝑎31 𝑎32 𝑎33 𝑎340001 coordinate vectors multiplied by the matrix define transformed coordinates
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu2. registrationFNIRT: non-linear transformationFMRIB’s Non-Linear Registration Tool inter-subject: same anatomical region in all subjects (atlas with anatomical labels) many many DOFs, same modality, least squares cost function, bias field model (FNIRT) represented by a 3D deformation field warp, displacement fieldstored as 3 x 3D volumes (x-, y-, z-components of the vectors)deformation nenthttps://fsl.fmrib.ox.ac.uk/fsl/fslwiki! inter-subject variability – not very accurate! some individual features losthttp://fsl.fmrib.ox.ac.uk/fslcourse-66 mm
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu2. registrationcost function registration iterative process of finding the best alignment of 3D volumes minimizing cost function measure of goodness of the alignmentlocal beware of local minima!min globalminlocalminleast squares: same modality, same brightness, contrast (includes bias field model)normalized correlation: same modality, different brightness, contrastcorrelation ratio: any MRI modalitiesmutual information: any modalities (also CT, PET etc.)normalized mutual information: any modalities (also CT, PET etc.)boundary-based-registration BBR: intra-subject, EPI to structural
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu2. /fslcourse finding intensity values between grid pointsnearest neighbor (NN)fast, blocky (pixelized)for labels, binary maskstri linear (TL)fast, blurringmost common optionspline, sincslower, sharper images(spline not that slow)7T Siemens GRE EPI 1.5 mm3
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduFLIRT: how to use it? flirt -in input –ref reference–out output -omat matrix-dof df-cost cf-interp itdegrees of freedom, 12cost function, corratiointerpolation method, trilinear-searchrx min max-nosearch-usesqform-noresamplesearch angles, -90 90all search angles 0initialize using sform/qformestimates transformation2. registration
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduFLIRT: example2. registration7T Siemens GRE EPI 1.5 mm37T Siemens MEMPRAGE 0.75 mm3 flirt -in input –ref reference –out output -omat matrix-cost mutualinfo -dof 6 –interp sinc–searchrx -3 3 –searchry -3 3 –searchrx -3 3
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu2. registrationFLIRT: mask registration applying existing transformation: flirt -in input –ref reference –out output–applyxfm –init matrix.mat-interp nearestneighbour-interp trilinearoriginalNNinteger, rounded mask shrinksfollowed by thresholding: 0.5 same size mask as input 0.5 more PVE, 0.5 less PVETLTL vs NNTL 0.5 vs NNTL 0.2 vs NN
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduFLIRT & FNIRT: weighted registration cost function weighting for input and/or reference imagevalues range [0,1], for corrupted areas, for important athologies 1 0-refweight volume -inweight volume use weights for reference volumeuse weights for input volume2. registration
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu2. registrationMCFLIRT: motion correction registration of multiple frames of the same fMRI scan mcflirt –in input –out output [options]-refvol reference frame, middle-meanvol use mean volume as a reference-cost cf-dof dfcost function, normcorrdegrees of freedom, 6before-sinc final-spline final final interpolation choice, trilinear-stats-mats-plotssave variance & stdev imagessave motion parameters in*.par filesave transformation matrices (in subdir)Jenkinson, M, et al., Improved Optimisation for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage, 17(2), 825-841, 2002.after
3. distortion correction
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu3. distortion correctionEPI geometric distortionmagnetic susceptibility 𝜒 material’s property, quantifies ability to magnetizesusceptibility effects T2*w contrast𝐵0 PA 𝚫𝝓 𝚫𝝓 𝚫𝝓brain χ 1 𝐵distortion𝝓𝝓𝝓0 𝚫𝝓 𝚫𝝓 𝚫𝝓air χ 0Gydephasing1. field map based distortion correction (FUGUE)2. EPI data acquired with the opposite PE direction (TOPUP)AP
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.eduFUGUE: how to use it?3. distortion correction field map B voxel shift map undo distortion (unwarp) need to have/know:field map, phase encoding direction, ESP, TEs ( TE) fugue -i epi –p phase –u output--unwarpdir dir–-dwell ESP--asym dTE-s sigmaphase encoding direction, y (usually AP)echo spacing (ESP, in sequence parameters) TE2D Gaussian smoothing (field map regularization)! phase data must have the same FOV & resolution as EPI (may require prior resampling)
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu3. distortion correctionFUGUE: how to use it?magnitude TE17T Siemens, 2mm3magnitude TE2 phase Siemens: GUI/script, magnitude data brain only (BET), output in rad/s fsl prepare fieldmap SIEMENS phz mag output dTE others (or Siemens) manually: resampling to EPI resolution: flirt -applyxfm convert phase to radians & rad/s: fslmaths unwrapping phase: prelude
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu {FSLDIR}/etc/flirtsch/TOPUP: distortion correction EPI data acquired with opposite phase encoding directions3. distortion correction
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu {FSLDIR}/etc/flirtsch/TOPUP: distortion correction EPI data acquired with opposite phase encoding directions3. distortion correction
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edu {FSLDIR}/etc/flirtsch/TOPUP: how to use it? 3. distortion correctiontopup -–imain input -–datain ap pa.txt –-config topup.cfg--out output --fout fmap-–iout corrected --dfout deform input: combined frames with opposite phase encoding directions ap pa.txt:columns 1-3 phase encoding directions (3rd one must be 0),column 4 readout time (if vary between the volumes) config: default one FSLDIR/etc/flirtsch/b02b0.cnf(data resolution 3.0 mm) corrected: input set of framesotherwise applytopupap pa.txt0 1 0 10 1 0 10 1 0 10 -1 0 10 -1 0 10 -1 0 1
Why N How Seminar Martinos Center March 30, 2017 Anna Blazejewska ablazejewska@mgh.harvard.edushell scriptingREF {PATH TO ALL MY DATA}/struct brain.nii.gzfor s in {PATH TO ALL MY DATA}/epi*brain.nii.gz; doflirt -in {s} –ref {REF} –dof 6 –cost mutualinfo.done; automatic! work by themselves!run multiple subjects/datasets automaticallysystematically explore different parameters/optionsrun multiple processing streams in parallelreproducibility: each subjects processed the same wayreplicability: easy to re-run with some modificationsave (are) documentation of what was runeasy to share with others! check the results on each stage of processing & monitor potential crashing.
Thank you!Anna Blazejewskaablazejewska@mgh.harvard.edu
head radius [mm], initial r/2 robust brain center estimation temporary add slices for small z-FOV apply to 4D fMRI data (-f 0.3 dilation)-R-Z-F-m-s-o-n-A save binary brain mask save skull image save brain surface outline skip segmented brain image generate skull & scalp surfaces (BETSURF)