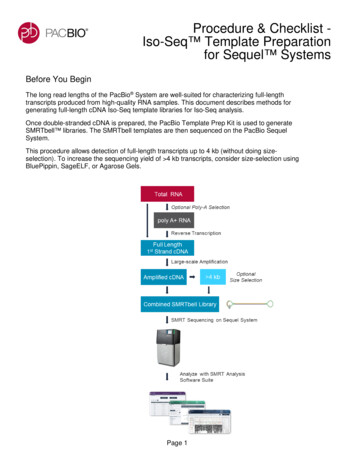
Transcription
Experimental ProtocolPlease note: the unsupported protocols described herein may not have been validated by Pacific Biosciences and are provided as-isand without any warranty. Use of these protocols is offered to those customers who understand and accept the associated terms andconditions and wish to take advantage of their potential to help prepare samples for analysis. If any of these protocols are to be used ina production environment, it is the responsibility of the end user to perform the required validation.Bacterial Iso-Seq Transcript SequencingUsing the SMARTer PCR cDNA Synthesis Kitand BluePippin Size-Selection SystemDescriptionThe long read lengths of the PacBio System are well-suited for charactering full-length transcripts produced fromhigh-quality RNA samples, including full-length operon transcripts from bacterial transcriptomes. This documentdescribes methods for generating full-length cDNA libraries (using the BluePippin System) for bacterial isoformsequencing.The reverse transcription kit utilized for this method, the SMARTer PCR cDNA Synthesis Kit from Clontech,requires the presence of a poly(A)-tail in order to generate full-length cDNA. Due to the absence of a poly(A)-tailon bacterial transcripts, users must first enzymatically add poly(A)-tails to 3’ ends of all transcripts (using thePoly(A) Polymerase Tailing Kit from Epicentre). Since this will add poly(A)-tails to all transcripts, including highlyabundant rRNA, users must then deplete rRNA using an rRNA-depletion method (RiboMinus Technology fromThermo Fisher is described in this protocol).Once double-stranded cDNA is prepared, the PacBio Template Prep Kit can be used to generate SMRTbell libraries. The SMRTbell templates are then sequenced on the PacBio System. Note that this protocol has beenoptimized for an input of 10 µg total RNA. It is very important to start with high quality RNA in order to obtainsequence coverage of full-length transcripts.Page 1
Experimental ProtocolRequired Reagents/ConsumablesItemVendorPoly(A) Polymerase Tailing KitEpicentre (PAP5104H)RiboMinus Transcriptome Isolation Kit, BacteriaThermo Fisher (K155004)RNeasy Mini KitQiagen (74104 or 74106)SMARTer PCR cDNA Synthesis KitClontech (634925 or 634926)KAPA HiFi PCR KitKapa Biosystems (KK2101 or KK2102)PrimeSTAR GXL DNA PolymeraseClontech (R050A or R050B)Additional 5’ PCR Primer IIAAny Oligo Synthesis Vendor1.2% FlashGel system or0.80% Agarose GelsFlashGel DNA Marker (100 bp - 4 kb)LonzaAny MLSLonzaNanodropThermo ScientificDNA 7500 Bioanalyzer KitDNA High Sensitivity Bioanalyzer KitRNA Pico Bioanalyzer Kit2100 Bioanalyzer SystemBluePippin system with Software v5.90 or laterPacBio SMRTbell cassette definition set “0.75% DF2 – 6kb Marker S1”0.75% Dye-Free Agarose Gel CassettesLoading SolutionS1 MarkerElectrophoresis BufferTemplate Prep KitDNA/Polymerase Binding KitMagBead KitDNA Sequencing KitAMPure PB BeadsDEPC-treated WaterNuclease-free WaterAgilentSage SciencePacific BiosciencesAny MLSPage 2
Experimental ProtocolWorkflowTotal RNAPolyA-TailrRNA Depletion1st Strand SynthesisPCR Cycle OptimizationLarge Scale PCRBluePippin System Size-SelectionLarge Scale PCRSMRTbell Template PreparationOptional 3 – 6 kb BluePippin System Size-SelectionPacBio RS IIPage 3
Experimental ProtocolPreparing RNA for Reverse TranscriptionPoly(A)-TailingPoly(A)-tails are added to 3’ ends of all transcripts by using the Epicentre Poly(A) Polymerase Tailing Kit. This kituses recombinant E. coli poly(A) polymerase to enzymatically add a poly(A)-tail that, if following the belowprocedure, will add a poly(A)-tail of approximately 200-300 nucleotides.The below procedure has been optimized for 10 µg of input total RNA. RNA concentration, reaction incubationlength, enzyme concentration and reaction volume all have an effect on poly(A)-tail length. If not enough startingmaterial is available, adjust these conditions to obtain a tail of 200-300 nucleotides (refer to Epicentre Poly(A)Polymerase Tailing Kit manual for more information).1. For each sample, combine the reagents below in separate sterile, RNase-free 0.5 mL LoBind tubes.This poly(A)-tailing has been optimized to add 200-300 nucleotide poly(A)-tails using 10 µg total RNA asinput. Do not change the reaction volumes unless less than 10 µg of starting material is available. Ifstarting with less than 10 µg total RNA, refer to Poly(A) Polymerase Tailing Kit manual to adjustreagent/reaction volumes and conditions accordingly.ReagentVolumePoly(A) Polymerase 10x Reaction Buffer5 μL10 mM ATP5 μLRNA (10 µg in 39 μL or less)NotesXto 50 μL total reactionvolume1 μLDEPC-Treated WaterPoly(A) Polymerase (4U/μL)50 μLTotal Volume2. Mix contents by pipetting and spin tubes briefly in a microfuge.3. Incubate the tubes at 37 C for 30 min.4. Purify reaction with Qiagen RNeasy Mini Kit per manufactures instructions. Elute in 30 μL DEPC-treatedwater.5. QC Steps: Measure concentration of samples using a Nanodrop instrument. Run a Bioanalyzer RNA Pico chip on the input RNA and the poly(A)-tailed RNA, making sure to diluteRNA samples accordingly. There should be a shift in the rRNA peaks of 200-300 nucleotides.Ribosomal RNA depletionIn order to prevent the majority of sequence coverage coming from highly abundant rRNA transcripts, these rRNAtranscripts must be removed from the RNA samples. This protocol describes the use of the RiboMinusTranscriptome Isolation Kit for Bacteria from Thermo Fisher. Oligo-based rRNA depletion kits are sequencesensitive, therefore one kit may not be suitable for all bacterial species. If the RiboMinus kit does not sufficientlyremove rRNA for a given sample, use another rRNA depletion kit (make sure the kit is specific for bacteria).This protocol has been optimized for an input of 2-10 µg of total RNA (RNA post poly(A)-tailing from previous step)in a volume of equal to or less than 20 μL.Page 4
Experimental ProtocolSTEP1234567Preparing RiboMinus Magnetic BeadsNotesResuspend the RiboMinus Magnetic Beads in its bottle by thoroughlyvortexing.For each sample, pipet 250 μL of the bead suspension into a sterile,RNase-free, 1.5 mL LoBind tube.Place the tube/s with the bead suspension on a magnetic rack for 1 minute.Gently aspirate and discard the supernatant.Add 250 μL DEPC-treated water to the beads and resuspend beads. Placethe tube/s on a magnetic rack for 1 minute. Gently aspirate and discard thesupernatant.Repeat Step 4 once.Resuspend beads in 250 μL Hybridization Buffer. Place tube/s on amagnetic rack for 1 minute. Gently aspirate and discard supernatant.Resuspend beads in 100 μL Hybridization Buffer and keep the beads at37 C until use.Selective HybridizationPerform hybridization of your total RNA sample with the RiboMinus Bacteria Probe.STEPHybridization1Per sample, to a sterile, RNase-free 1.5 mL tube, add:Total RNA (2-10 μg): 20 μL (remaining 10 μL of poly(A)-tailed totalRNA should be stored at -80 C)RiboMinus Probe (100 pmol/μl): 4 μlHybridization Buffer: 100 μl2Incubate the tube/s at 37 C for 5 minutes to denature RNA34Place the sample/s on ice for at least 30 seconds.5678910Briefly centrifuge the tube/s with the cooled hybridized sample (from Step3) to collect the sample to the bottom of the tube.Transfer the sample/s ( 124 µL) to the prepared RiboMinus Magneticbeads.Mix well by vortexing the tube repeatedly.Incubate the tube/s at 37 C for 15 minutes. During incubation, gently mixthe contents occasionally.After incubation, place the tube/s on a magnetic rack for 1 minute to pelletthe rRNA-probe complex. Do not discard the supernatant. Thesupernatant contains RiboMinus RNA.Transfer the supernatant ( 224 µL) to a sterile, RNase-free 1.5 mL tube.Clean-up and concentrate 100 μL of rRNA depleted RNA with QiagenRNeasy Mini Kit per manufactures’ instructions. Elute in 30 μL DEPCtreated water. Store remaining 124 μL of rRNA depleted RNA at -80 C,for use in other experiments. Poly(A)-tailed, rRNA depleted RNA can beused as input material for cDNA generation.Page 5Notes
Experimental ProtocolPreparing cDNA from RNA SamplesFirst-Strand SynthesisFirst-strand cDNA synthesis employs the SMARTer PCR cDNA Synthesis Kit. The CDS Primer IIA is firstannealed to the polyA tail of transcripts, followed by first-strand synthesis with SMARTScribe ReverseTranscriptase. The first-strand product is diluted with Elution Buffer (EB) to an appropriate volume andsubsequently used for large-scale PCR.1. For each sample, combine the reagents below in separate sterile, RNase-free 0.5 mL LoBind tubes tocreate first-strand reaction tubes. For bacterial Iso-Seq transcript sequencing, we recommend making 4 first-strand reactions per sample.Do not change the size (volumes) of any of the reactions. All components have been optimized for thevolumes specified.ReagentVolumeNotes3.5 μLPoly(A), rRNA depleted RNA3’ SMART CDS Primer II A (12 μM)1 μLNuclease-Free WaterX4.5 μLTotal Volume2. Mix contents and spin the tubes briefly in a microfuge.3. Incubate the tubes at 72 C in a hot -lid thermal cycler for 3 minutes; slow ramp to 42 C at 0.1 C/sec andthen let sit for 2 minutes.During this incubations step, prepare a master mix for all 4 first-strand reactions prepared above, atroom temperature, by combining the following reagents in the order shown. It is important to goimmediately into step 4 after step 3. However, add the reverse transcriptase to the master mix just priorto use. Mix well by pipetting and spin the tube briefly in a microfuge.ReagentVolume2 μL5X First-Strand BufferDTT (100 mM)0.25 μLdNTP (10 mM)1 μLSMARTer II A Oligonucleotide (12 μM)1 μL0.25 μLRNase InhibitorSMARTScribe Reverse Transcriptase (100 U) - add before use5.6.7.1 μL5.5 μLTotal Volume added per reaction4.NotesAfter adding reverse transcriptase, place the master mix at 42 C for 1 minute to bring it up to temperatureand proceed immediately to step 4.For this step, make sure thermal cycler stays heated to 42 C. While first -strand reaction tubes are still onthermal cycler block, aliquot 5.5 μL of the Master Mix into each first-strand reaction tube. Mix thecontents of the tubes by gently pipetting, and spin the tubes briefly to collect the contents at the bottom.Immediately return tubes to thermal cycler.Incubate the tubes at 42 C for 90 minutes.Terminate the reaction by heating the tubes at 70 C for 10 minutes.Dilute each first-strand reaction with 40 μL of PacBio Elution Buffer (EB) and pool diluted first-strandreactions for the same RNA sample.Page 6
Experimental ProtocolLarge-Scale PCRIt is highly recommended to perform cycle optimization to determine the optimal number of cycles (whileminimizing artifacts during large-scale amplification) for large-scale PCR.PCR Cycle OptimizationIn this section, perform test amplifications to determine the best number of cycles required for the sample.Collect a total of 5, 5 μL aliquots from recommended cycle below.1. Add the following reagents to an appropriately sized PCR tube:ReagentVolumeKAPA HiFi Fidelity Buffer (5X)10 μLDiluted first-strand cDNA from step 7 above10 μLKAPA dNTP Mix (10 mM)1.5 μL5’ PCR Primer II A (12 μM)3.2 μLNuclease-free water24.3 μLKAPA HiFi Enzyme (1 U/μL)1 μLTotal Volume50 μLNotes2. Cycle the reaction with the following conditions (using a heated lid):3. After the initial 8 cycles, remove 5 μL of the reaction and transfer it to a tube labeled “8”.4. Return the remaining 45 μL PCR reaction to the thermal cycler and run 2 additional cycles as below: 2 cycles at the following temperatures and times:o 98 C for 20 secondso 65 C for 15 secondso 72 C for 4 minutes Final extension:o 72 C for 5 minutes5. Remove 5 μL again and transfer to a tube labeled “8”.6. Repeat steps 4-5 for 10, 12, 14, 16 and 18 cycles.Note that the number of cycles is dependent on the sample, and may be changed for particular samples.Therefore, it may be necessary to adjust which cycle numbers are used for optimization.7. Load the 6 aliquots on an agarose gel or Lonza flash gel to view the distribution of the ds-cDNA.See UB – Guidelines for Preparing cDNA Libraries for Isoform Sequencing (Iso-Seq Analysis).Page 7
Experimental ProtocolLarge-Scale PCR for Size-Selection on the BluePippin SystemUse the cycle number (as determined in the PCR Cycle Optimization step) to generate ds-cDNA forsize-selection on the BluePippin system.1. Set up 8 x 50 μL PCR reactions.2. Make a master mix by adding the following reagents:ReagentVolumeKAPA HiFi Fidelity Buffer (5X)80 μLDiluted first-strand cDNA Synthesis80 μLKAPA dNTP Mix (10 mM)12 μL5’ PCR Primer II A (12 μM)25.6 μLNuclease-free water194.4 μLKAPA HiFi Enzyme (1U/μL)Notes8 μL400 μLTotal Volume3. Transfer 50 μL aliquots into 8 PCR tubes and perform PCR using the cycle number determinedduring the optimization step. Cycle the reaction with the following conditions (using a heated lid): Initial denaturationo95 C for 2 minutes n cycles (optimal cycle determined in the optimization step) at the following temperatures and times:o 98 C for 20 secondso 65 C for 15 secondso 72 C for 4 minutes Final extension:o 72 C for 5 minutesPage 8
Experimental ProtocolSTEPPool 8 PCR Reactions and Purify1Add 1.0X volume of AMPure PB magnetic beads.23Mix the bead/DNA solution thoroughly.4Allow the DNA to bind to beads by shaking in a VWR vortex mixer at 2000 rpmfor 10 minutes at room temperature.5Spin down the (for 1 second) to collect beads.6Place the tube in a magnetic rack until the beads collect to the side of thetube.7With the tube still on the magnetic rack, slowly pipette off cleared supernatantand save in another tube. Avoid disturbing the bead pellet.8Wash beads with freshly prepared 70% ethanol.9Repeat step 8 above.10Remove residual 70% ethanol. Remove the tube for magnetic rack and spin to pellet beads. Both thebeads and any residual 70% ethanol will be at the bottom of the tube. Place the tube back on magnetic rack. Pipette off any remaining 70% ethanol.11Check for any remaining droplets in the tube. If droplets are present, repeatstep 10.Quickly spin down the tube (for 1 second) to collect the beads. Do not pelletbeads.Remove the tube from the magnetic rack and allow beads to air-dry (with thetube caps open) for 30 to 60 seconds.Elute the DNA off the beads in 61 μL of Elution Buffer. Vortex for 10 minutesat 2000 RPM. Measure sample concentration on the Nanodrop instrument with 1 μL ofsample. Save 1 μL sample used for the Nanodrop instrument to run on theBioanalyzer instrument.Page 9Notes
Experimental ProtocolSize-Selection using the BluePippin SystemSTEPSize-Selection1Follow the BluePippin manual and instructions to calibrate your instrument. A new calibration is required before each BluePippin run.2Inspect the gel cassette (using Sage Sciences’s BluePippin manual). Ensure that the buffer wells are full. Ensure that there is no separation of the gel from the cassette.3Prepare the gel cassette: Remove all bubbles from the elution buffer chamber by tilting the cassetteand tapping it until all air bubbles move into the buffer chamber. Place the gel cassette in the BluePippin System and carefully remove theplastic seals on the cassette. Remove the buffer from the elution well and fill with 40 μL of freshElectrophoresis Buffer. Keep the pipette down the center of the well and avoid creating a vacuumin the well. The bottom of the well is okay to touch. If the well “bubbles” over when adding the buffer to the well, remove bufferand try again. If the well continues to “bubble” over, then use this well forthe S1 marker. The elution well may be damaged and should not be usedfor sample collection. Cover the elution wells with a clear adhesive tape Remove the buffer from the sample well and fill with 70 μL of freshElectrophoresis Buffer Close the lid and perform a Continuity Test.4Prepare samples for loading onto the BluePippin: For fraction 1 - 2kb, prepare an aliquot of 500-750 ng in 30 μL. For fraction 2 - 3kb, prepare an aliquot of 1-1.5 μg in 30 μL. For fraction 3 - 6kb, prepare an aliquot of 2-5 μg in 30 μL. Use Elution Buffer to dilute samples to 30 μL. Add 10 μL of the BluePippin Loading Solution and mix well by pipetting,avoid creating bubbles. Briefly spin down samples in microfuge.5Loading Samples: Remove 40 μL of buffer from each well. Load all 40 μL of the sample prepared in step 4 into each lane. Load 40 μL of S1 Marker in one of the lanes.Page 10Notes
Experimental Protocol6Sep up the run protocol: Click on the “New” button to create a new protocol. Select the “0.75% DF 2 – 6kb Marker S1” cassette definition file. Click on the box below “End Run when Elution is Complete.” Set the lane where the S1 Marker is loaded as the reference “Ref” lane. Click on the “Range” button and enter the following:Library SizeBP StartBP End1 kb - 2 kb50020002 kb - 3 kb150030003 kb - 6 kb250060007Start the run.89After the run, collect approximately 40 μL from the elution well of each lanerun with a sample. After collecting all fractions, pipette 40 μL Elution Buffer into the elutionwell of all lanes run with a sample. One by one, mix the Elution Buffer in the elution well by pipetting gently,avoiding bubbles. After mixing, collect this second 40 μL and combine it with the first elutionfor each respective sample. There is now a total of 80 μL of sample per size fraction.Quantification and purification are not necessary at this point. A HighSensitivity Bioanalyzer instrument can be run to determine effectiveness ofsize-selection.Page 11
Experimental ProtocolLarge-Scale PCR for SMRTbell Library PreparationAfter size selection, the double-stranded cDNA is not sufficient for SMRTbell library construction. Performlarge-scale PCR using the eluted DNA from the previous step to generate more double-stranded cDNA.Note for the post BluePippin large-scale PCR, PrimeSTAR GXL DNA Polymerase from Clontech is used foramplification.PCR for Each Fraction1. Set up 8 X 50 μL PCR reactions for each size fraction: 1 kb – 2 kb, 2 kb – 3 kb and 3 kb – 6 kb.2. Add the following reagents to an appropriately sized master mix tube:ReagentVolumePrimeSTAR GXL Buffer (5x)80 μLdNTP Mix (10 mM)32 μLEluted DNA from the BluePippin40 μL5’ PCR Primer II A (12 μM)20 μLNuclease-free water220 μLPrimeSTAR GXL Enzyme (1.25U/μL)Notes8 μL400 μLTotal Volume3. Aliquot 50 μL into 8 PCR tubes and perform PCR using the cycle number and extension parametersbelow:Size DesiredExtension TimeNumber of Cycles1 kb - 2 kb1 min10 cycles2 kb - 3 kb2 min12 cycles3 kb - 6 kb4 min14 cycles4. Cycle the reactions with the following conditions (using a heated lid): Initial denaturation:o 98 C for 30 seconds n cycles at the following temperatures and times:o 98 C for 10 secondso 65 C for 30 secondso 68 C for x minutes (for this step, see extension times in table above) Final extension:o 68 C for 5 minutes5. Pool reactions into a 1.5 mL tube and purify the reactions for each size fraction using AMPure PB beads aspreviously described with the following fold-amounts of AMPure PB beads per size-bin: 1 – 2 kb use 1.0X 2 – 3 kb use 0.6X 3 – 6 kb use 0.6X6. Elute each reaction in 31 µL Elution Buffer.7. Check concentration of each sample on a Nanodrop instrument.8. Check sample size distribution buy running each sample on a Bioanalyzer 7500 chip.9. If there are small fragments present in the 3 – 6 kb size-bin, it may be necessary to run an additionalBluePippin size-selection at the end of the template prep procedure to avoid preferentially loading theseshort fragments during sequencing.Page 12
Experimental ProtocolcDNA SMRTbell Template Preparation and SequencingRepair DNA DamageIn general, 1 – 1.5 μg of cDNA is recommended for each sample going into SMRTbell template preparation. Forsize-selected samples, the input amount depends on the size:If preparing larger amounts of DNA than what the table below indicates, scale the reaction volumes accordingly.FractionInput Requirement1 kb - 2 kbUp to 500 ng2 kb - 3 kbUp to 1 μg3 kb -
SMARTScribe Reverse Transcriptase (100 U) - add beforeuse 1 μL Total Volume added per reaction 5.5 μL After adding reverse transcriptase, place the master mix at 42 C for 1 minute to bring it up to temperature and proceed immediately to step 4. 4. For this step, make sure thermal cycler stays heated to File Size: 305KB