Transcription
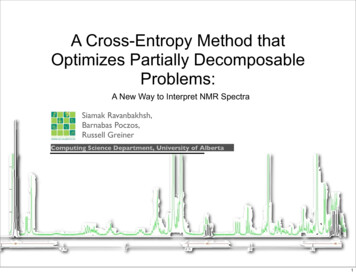
A Cross-Entropy Method thatOptimizes Partially DecomposableProblems:A New Way to Interpret NMR SpectraSiamak Ravanbakhsh,Barnabas Poczos,Russell GreinerComputing Science Department, University of Alberta1
Metabolomics & NMR SpectroscopyMetabolomics study of chemical fingerprints that cellular processes leavebehindComputing Science Department, University of AlbertaAAAI’102
Metabolomics & NMR SpectroscopyMetabolomics study of chemical fingerprints that cellular processes leavebehindMetabolites: end products ofgene expressionComputing Science Department, University of AlbertaAAAI’102
Metabolomics & NMR SpectroscopyMetabolomics study of chemical fingerprints that cellular processes leavebehindMetabolites: end products ofgene expression{Gas chromatographyHigh performance liquid chromatographyCapillary electrophoresisMass Spectrometrysource: wikipediaToolbox:Nuclear Magnetic Resonance (NMR) SpectroscopyComputing Science Department, University of AlbertaAAAI’102
Nuclear Magnetic Resonance (NMR) SpectroscopyComputing Science Department, University of AlbertaAAAI’103
Nuclear Magnetic Resonance (NMR) SpectroscopyNMR SpectrumComputing Science Department, University of AlbertaAAAI’103
Nuclear Magnetic Resonance (NMR) Spectroscopyheight ( a )width (w)Spectrum is made of manyLorentzian peaksoffset ( )NMR SpectrumComputing Science Department, University of AlbertaAAAI’103
3-Hydroxyisobutyric acidComputing Science Department, University of AlbertaAAAI’104
3-Hydroxyisobutyric acidComputing Science Department, University of AlbertaAAAI’104
clusterShift3-Hydroxyisobutyric acidComputing Science Department, University of AlbertaAAAI’104
ConcentrationclusterShift3-Hydroxyisobutyric acidComputing Science Department, University of AlbertaAAAI’104
The Goal is:Given The library of metabolite SignaturesFind Corresponding ConcentrationComputing Science Department, University of AlbertaAAAI’105
The Goal is:Given The library of metabolite SignaturesFind Corresponding ConcentrationOptimization variables: Metabolite Concentrations Chemical ShiftsComputing Science Department, University of AlbertaAAAI’105
The Goal is:Given The library of metabolite SignaturesFind Corresponding ConcentrationOptimization variables: Metabolite Concentrations Chemical ShiftsIt’s a difficult Problem Nonlinear in shift variables Involves hundreds of variables Loss is very expensive to evaluate (100K points) Non-Convex even around local optima Incomplete library results in over-fittingComputing Science Department, University of AlbertaAAAI’105
Exploiting the structureComputing Science Department, University of AlbertaAAAI’106
Exploiting the structureSub-problemsare easier to solvebut theyshare variablesComputing Science Department, University of AlbertaAAAI’106
Exploiting the structureSub-problemsare easier to solvebut theyshare variablesComputing Science Department, University of AlbertaAAAI’106
Coupling Matrixrepresents the structureComputing Science Department, University of AlbertaAAAI’107
Coupling Matrixrepresents the structureSub-problemsVariablesComputing Science Department, University of AlbertaAAAI’107
Coupling Matrixrepresents the structureVariablesSub-problemsSudoku PuzzleComputing Science Department, University of AlbertaAAAI’107
Coupling Matrixrepresents the structureVariablesSATisfiability ProblemComputing Science Department, University of AlbertaAAAI’107
Cross Entropy (CE)Method for OptimizationComputing Science Department, University of AlbertaAAAI’108
Cross Entropy (CE)Method for OptimizationStart from a priorLoss functionPriorComputing Science Department, University of AlbertaAAAI’108
Cross Entropy (CE)Method for OptimizationStart from a priorLoss functionRepeat until convergenceTake samples from current dist.Calculate the loss for samplesPriorComputing Science Department, University of AlbertaAAAI’108
Cross Entropy (CE)Method for OptimizationStart from a priorLoss functionRepeat until convergenceTake samples from current dist.Calculate the loss for samplesSelect Elite samplesPriorComputing Science Department, University of AlbertaAAAI’108
Cross Entropy (CE)Method for OptimizationStart from a priorRepeat until convergenceTake samples from current dist.Calculate the loss for samplesSelect Elite samplesFind maximum likelihood dist. for ElitesComputing Science Department, University of AlbertaAAAI’108
Cross Entropy (CE)Method for OptimizationStart from a priorRepeat until convergenceTake samples from current dist.Calculate the loss for samplesSelect Elite samplesFind maximum likelihood dist. for ElitesA subroutine to be used againComputing Science Department, University of AlbertaAAAI’108
Variation of CEVariables ( i )Sub-problems (k)that Exploits Decomposability (CEED)coupling matrixComputing Science Department, University of AlbertaAAAI’109
Variation of CEVariables ( i )Start from a priorRepeat,Sub-problems (k)that Exploits Decomposability (CEED)coupling matrixUntil convergenceComputing Science Department, University of AlbertaAAAI’109
Variation of CEVariables ( i )Start from a priorRepeat,For each sub-problem kSub-problems (k)that Exploits Decomposability (CEED)Draw samples from marginal of current dist.Calculate the loss for samplescoupling matrixSelect Elite samplesFind maximum likelihood dist. for ElitesUntil convergenceComputing Science Department, University of AlbertaAAAI’109
Variation of CEVariables ( i )Start from a priorRepeat,For each sub-problem kSub-problems (k)that Exploits Decomposability (CEED)Draw samples from marginal of current dist.Calculate the loss for samplescoupling matrixSelect Elite samplesFind maximum likelihood dist. for ElitesFor each variable iCombinedist’s from related sub-problemsUntil convergenceComputing Science Department, University of AlbertaAAAI’109
How toCombineML DistributionsComputing Science Department, University of AlbertaAAAI’1010
How toML DistributionsWe use linear combination by Fisher InformationCombineComputing Science Department, University of AlbertaAAAI’1010
How toML DistributionsWe use linear combination by Fisher InformationCombineSecond distributionCombined distributionGaussian DistributionsFirst distributionComputing Science Department, University of AlbertaAAAI’1010
How toML DistributionsWe use linear combination by Fisher InformationCombineGaussian DistributionsComputing Science Department, University of AlbertaAAAI’1010
How toML DistributionsWe use linear combination by Fisher InformationCombineBeta DistributionsComputing Science Department, University of AlbertaAAAI’1010
How toML DistributionsWe use linear combination by Fisher InformationCombineBeta DistributionsComputing Science Department, University of AlbertaAAAI’1010
How toML DistributionsWe use linear combination by Fisher InformationCombineBernoulli DistributionsComputing Science Department, University of AlbertaAAAI’1010
How toML DistributionsWe use linear combination by Fisher InformationCombineBernoulli DistributionsA Tweak for NMR problemComputing Science Department, University of AlbertaAAAI’1010
Experimental Results800 MHz SpectraComputing Science Department, University of AlbertaAAAI’1011
Experimental Results500 MHz SpectraComputing Science Department, University of AlbertaAAAI’1011
Experimental ResultsSimulated SpectraComputing Science Department, University of AlbertaAAAI’1011
Experimental ResultsComparison with ChenomX Inc. automated fitting softwareQuantification TaskMetabolite Detection TaskThreshold: .02 mMolAvg. Relative ErrorUsThemComputing Science Department, University of AlbertaAAAI’1011
Experimental ResultsComputing Science Department, University of AlbertaAAAI’1011
MaxSATAnalytically update of dist’s is possible.variableConvergence of distributions to the correct assignmentiterationComputing Science Department, University of AlbertaAAAI’1012
MaxSAT#clauses 4250#vars1000# satisfied clausesConvergence of CE and CEEDiterationComputing Science Department, University of AlbertaAAAI’1012
Sudokusolution in redUsing categorical distributionOur Alg. is 5 times faster than CEComputing Science Department, University of AlbertaAAAI’1013
ConclusionOur method successfully exploits partial decomposability intargeted profiling of NMR spectraas well as some combinatorial problems (i.e. SAT & Sudoku)Maximum likelihood estimates could be linearly combined based ontheir certainty using their Fisher Information.Computing Science Department, University of AlbertaAAAI’1014
Thank you!Computing Science Dept. University of AlbertaAlberta Ingenuity Centre for Machine Learning15
that Exploits Decomposability (CEED) coupling matrix Draw samples from marginal of current dist. For each sub-problem k Calculate the loss for samples Select Elite samples Find maximum likelihood dist. for Elites 9. Computing Science Department, University of Alberta AAAIÕ10 Start from a prior