Transcription
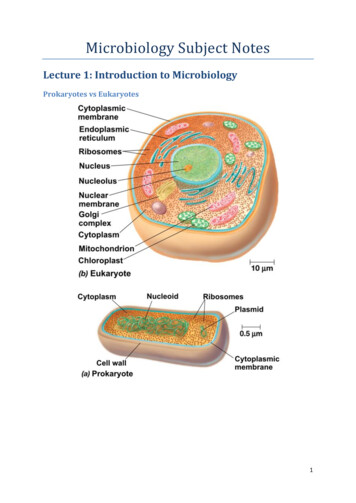
Microbiology Subject NotesLecture 1: Introduction to MicrobiologyProkaryotes vs Eukaryotes1
Infectious DiseaseBiofilms A biofilm is any group of microorganisms in which cells stick to each other on a surfaceThese adherent cells are frequently embedded within a self-produced matrix of extracellularpolymeric substance (EPS)In nature microorganisms typically live in communities (Biofilms), not as single speciescolonies as in the laboratory platesBiofilms improve survival by trapping nutrients, protecting against the environment, andallowing for favourable metabolic conditionsMicroorganisms and Agriculture Positive impactso Nitrogen-fixing bacteriao Cellulose-degrading microbes in the rumeno Regeneration of nutrients in soil and watero Dairy products (e.g., cheeses, yogurt, buttermilk)o Other food products (e.g., sauerkraut, pickles, leavened breads, beer)o The role of microbes in biofuels productionNegative impactso Diseases in plants and animalso Food spoilage by microorganisms2
Metabolic Diversity Chemoorganotrophs: Obtain their energy from the oxidation of organic moleculesChemolithotrophs: Obtain their energy from the oxidation of inorganic molecules, aprocess found only in prokaryotesPhototrophs: Contain pigments that allow them to use light as an energy sourceAutotrophs: Use carbon dioxide as their carbon source, sometimes referred to asprimary producersHeterotrophs: Require one or more organic molecules for their carbon source,feed directly on autotrophs or live off products produced by autotrophsLecture 2: Microbial Anatomy, Nutrition, and GrowthCell Morphology Major cell morphologieso Coccus (pl. cocci): spherical or ovoido Rod: cylindrical shapeo Spirillum: spiral shapeCells with unusual shapeso Spirocheteso Appendaged bacteriao Filamentous bacteria3
Determining Cell Shape Morphology typically does not predict physiology, ecology, phylogeny, etc of a cellSelective forces may be involved in setting the morphology, such as:o Optimization for nutrient uptake (those with high surface-to-volume ratio)o Swimming motility in viscous environments (helical or spiral-shaped cells)o Gliding motility (filamentous bacteria)Cell Sizeo Size range for prokaryotes: 0.2 µm to 700 µm in diametero Most cultured rod bacteria are between 0.5 and 4.0 µm wide and 15 µm longo Size range for eukaryotic cells: 10 to 200 µm in diameterThe Cytoplasmic Membrane in Bacteria The membrane is a thin structure that surrounds the cell, about 6–8 nm thick consisting of aphospholipid bilayer with integrated proteins and carbohydratesIt is a highly selective permeable barrier; enables concentration of specific metabolites andexcretion of waste productsThe outer surface of cytoplasmic membrane can interact with a variety of proteins that bindsubstrates or process large molecules for transportThe inner surface of cytoplasmic membrane interacts with proteins involved in energyyielding reactionsand other important cellular functionsIntegral membrane proteins are firmly embedded in the membrane, while peripheralmembrane proteins have one portion anchored in the membrane4
Transport Proteins Three major classes of active transport systems in prokaryoteso Simple transporto Group translocationo ABC systemo All require energy in some form, usually proton motive force or ATP Simple Transport example: Lac Permease of Escherichia coli, where lactose is transportedinto E. coli by the simple transporter lac permease, a symporter5
Group Translocation example: Phosphotransferase System in E. coli, where the substancetransported is chemically modified during transport across the membrane. Used to moveglucose, fructose, and mannose into the cells. Five proteins required ABC (ATP-Binding Cassette) transport Systems: transmembrane proteins that utilize theenergy of adenosine triphosphate (ATP) binding and hydrolysis to carry out certain biologicalprocesses including translocation of various substrates across membranes. Often involved inuptake of organic compounds (e.g., sugars, amino acids), inorganic nutrients (e.g., sulfate,phosphate), and trace metals6
Translocases: a general term for a protein that assists in moving another molecule, usuallyacross a membrane. For example, the sec translocase system exports proteins and insertsintegral membrane proteins into the membraneCell Walls Peptidoglycan: a polymer consisting of sugars and amino acids that forms a mesh-like layeroutside the plasma membrane of most bacteria, forming the cell wallThe sugar component consists of alternating residues of N-acetylglucosamine and Nacetylmuramic acid. Attached to the N-acetylmuramic acid is a peptide chain of three to fiveamino acids. The peptide chain can be cross-linked to the peptide chain of another strandforming the 3D mesh-like layerStructural differences between cell walls of gram-positive and gram-negative Bacteria areresponsible for differences in the Gram stain reactionThe peptidoglycan layer is substantially thicker in Gram-positive bacteria (20 to 80nanometers) than in Gram-negative bacteria (7 to 8 nanometers). Thus, presence of high7
levels of peptidoglycan is the primary determinant of the characterisation of bacteria asgram-positive Gram positive: Can contain up to 90% peptidoglycan, common to have teichoic acids (acidicsubstances) embedded in the cell wall Gram negative: a class of bacteria that do not retain the crystal violet stain used in the Gramstaining method of bacterial differentiation. They have a thin peptidoglycan layer, an Outermembrane containing lipopolysaccharide (LPS), porins in the outer membrane which act likepores for particular molecules, and a space between the peptidoglycan layer and thesecondary cell membrane called the periplasmic space. No teichoic acids are present8
Mycoplasmas: a genus of bacteria that lack a cell wall around their cell membrane. Withouta cell wall, they are unaffected by many common antibiotics such as penicillin or other betalactam antibiotics that target cell wall synthesisCell Surface Structures Capsules and Slime Layers: polysaccharide layers surrounding the cell wall or membrane thatassist in attachment to surfaces, protect against phagocytosis, and help resist desiccation.Mutant strains of S. pneumoniae that have lost the ability to form a capsule are readily takenup by white blood cells and do not cause diseaseFimbriae: an appendage composed of curlin proteins that can be found on many Gramnegative and some Gram-positive bacteria that is thinner and shorter than a flagellum.Fimbriae are used by bacteria to adhere to one another and to adhere to animal cells andsome inanimate objectsPili: filamentous protein structures typically longer than fimbriae, which assist in surfaceattachment and facilitate genetic exchange between cells (conjugation)Endospores An endospore is a dormant, tough, and non-reproductive structure produced by certainbacteria from the Firmicute phylumThe name "endospore" is suggestive of a spore or seed-like form (endo means within), but itis not a true spore (i.e., not an offspring). It is a stripped-down, dormant form to which thebacterium can reduce itselfEndospore formation is usually triggered by a lack of nutrients, and usually occurs in Grampositive bacteria9
Endospores enable bacteria to lie dormant for extended periods, even centuries. Revival ofspores millions of years old has been claimedEndospores can survive without nutrients. They are resistant to ultraviolet radiation,desiccation, high temperature, extreme freezing and chemical disinfectantsUp to 20% of the dry weight of the endospore consists of calcium dipicolinate within thecore, which is thought to stabilize the DNAThe core contains the spore chromosomal DNA which is encased in chromatin-like proteinsknown as SASPs (small acid-soluble spore proteins), that protect the spore DNA from UVradiation and heatFlagella and Motility A flagellum is a lash-like appendage that protrudes from the cell body of certain prokaryoticand eukaryotic cells. The word flagellum in Latin means whipThe primary role of the flagellum is locomotion but it also often has function as a sensoryorganelleThe bacterial flagellum is made up of the protein flagellin. Its shape is a 20 nanometer-thickhollow tube. It is helical and has a sharp bend just outside the outer membrane; this "hook"allows the axis of the helix to point directly away from the cellSeveral genes are required for flagellar synthesis and motility. It is synthesised progressively,from the base upwards10
Gliding Motility Gliding motility is the non-flagellar movement of bacteria on surfaces.Slower and smoother than swimmingMovement typically occurs along long axis of cellRequires surface contact, and works by excretion of polysaccharide slime and gliding-specificproteins11
Microbial Taxes Taxis: directed movement in response to chemical or physical gradientsChemotaxis: Bacteria respond to temporal difference in chemical concentration, withattractants and receptors sensed by chemoreceptorsPhototaxis: response to lightAerotaxis: response to oxygenOsmotaxis: response to ionic strengthHydrotaxis: response to waterMeasured by inserting a capillary tube containing an attractant or a repellent in a medium ofmotile bacteriaLecture 3: Microbial Genetics12
DNA StructureDNA Supercoiling DNA supercoiling refers to the over- or under-winding of a DNA strand, and is an expressionof the strain on that strand. Supercoiling is important in a number of biological processes,such as compacting DNASupercoiling of DNA reduces the space and allows for much more DNA to be packaged withthe cellSupercoiling is also required for DNA/RNA synthesis. The region ahead of the polymerasecomplex will be unwound; this stress is compensated with positive supercoils ahead of the13
complex. Behind the complex, DNA is rewound and there will be compensatory negativesupercoilsTopoisomerases, enzymes which regulation under or overwinding of DNA, such as DNAgyrase (Type II Topoisomerase) play a role in relieving some of the stress during DNA/RNAsynthesis by forming negative supercoilsNegative supercoiling: double helix is underwound. (predominantly found in nature)Positive supercoiling: double helix is overwoundTypes of Genetic Material Chromosome: a genetic element with “housekeeping” genes. The presence of essentialgenes is necessary for a genetic element to be called a chromosomePlasmid: a genetic element that is expendable and rarely contains genes for growth under allconditions. Usually circular and contain beneficial characteristics, like antibiotic resistance14
Transposable Elements: segment of DNA that can move from one site to another site on thesame or a different DNA molecule. Three main types: insertion sequences, transposons,special virusesEscherichia coli Chromosome Escherichia coli is a useful model organism for the study of biochemistry, genetics, andbacterial physiologyMany genes encoding enzymes of a single biochemical pathway are clustered into operonsThe Biology of Plasmids A cell can contain more than one plasmid, but it cannot be closely related genetically due toplasmid incompatibilityPlasmids belonging to same Inc group exclude each other from replicating in the same cellbut can coexist with plasmids from other groupsSome plasmids (episomes) can integrate into the cell chromosome; similar to situation seenwith prophagesConjugative plasmids can be transferred between suitable organisms via cell-to-cell contactGenetic information encoded on plasmids is not essential for cell function under allconditions but may confer a selective growth advantage under certain conditions15
R plasmids are resistance plasmids; confer resistance to antibiotics and other growthinhibitors; many are conjugativeIn several pathogenic bacteria, virulence characteristics are encoded by plasmid genesBacteriocins: proteins produced by bacteria that inhibit or kill closely related species or evendifferent strains of the same speciesDNA Replication DNA synthesis is bidirectional in prokaryotes: two replication forks moving in oppositedirectionsReplisome: complex of multiple proteins involved in replicationMutation rates in cells are 10 -8 to 10 -11 errors per base insertedPolymerase can detect mismatch through incorrect hydrogen bonding16
Polymerase Chain Reaction The polymerase chain reaction (PCR) is a technology in molecular biology used to amplify asingle copy or a few copies of a piece of DNA across several orders of magnitude, generatingthousands to millions of copies of a particular DNA sequenceDeveloped in 1983 by Kary Mullis, PCR is now a common and often indispensable techniqueused in medical and biological research labs for a variety of applicationsSteps:o Add DNAo Add primero Add DNA polymeraseo Heat and cool (repeated)The method relies on thermal cycling, consisting of cycles of repeated heating and cooling ofthe reaction for DNA melting and enzymatic replication of the DNA. Primers (short DNAfragments) containing sequences complementary to the target region along with a DNApolymerase, are key components to enable selective and repeated amplificationAs PCR progresses, the DNA generated is itself used as a template for replication, setting inmotion a chain reaction in which the DNA template is exponentially amplifiedRNA Transcription Transcription (DNA to RNA) is carried out by RNA polymerase from 5' to 3'Only one of the two strands of DNA are transcribed by RNA polymerase for any genePromoters: site of initiation of transcription, recognized by sigma factor of RNA polymeraseTranscription stops at specific sites called transcription terminatorsMost genes encode proteins, but some RNAs are not translated (i.e., rRNA, tRNA). There arethree types of rRNA: 16S, 23S, and 5S. rRNA and tRNA are very stable, while mRNAs haveshort half-lives (a few minutes)Prokaryotes often have genes clustered together to be transcribed all at once as a singlepolycistronic mRNA. An operon is a group of related genes cotranscribed on a polycistronicmRNA17
Protein Structure and Synthesis Start codon: translation begins with AUGStop codons: terminate translation (UAA, UAG, and UGA)Open reading frame (ORF): AUG followed by a number of codons and a stop codon in thesame reading frameA transfer RNA is a molecule composed of RNA, typically 76 to 90 nucleotides in length, thatserves as the physical link between the nucleotide sequence of nucleic acids (DNA and RNA)and the amino acid sequence of proteinstRNA and amino acid brought together by aminoacyl-tRNA synthetasesAnticodon: three bases of tRNA that recognize three complementary bases on mRNARibosomes: sites of protein synthesis, thousands per cell, composed of two subunits (30Sand 50S in prokaryotes)Translation is broken down into three main steps: Initiation, where two ribosomal subunitsassemble with mRNA beginning at AUG; Elongation, where amino acids are brought to theribosome and are added to the growing polypeptid; and Termination, when ribosomereaches a stop codon and Release factors (RF) recognize stop codon and cleave polypeptidefrom tRNAPolysomes: a complex formed by ribosomes simultaneously translating a single mRNA18
19
Mutations and Mutants Mutation: Heritable change in DNA sequence that can lead to a change in phenotype(observable properties of an organism)Mutant: A strain of any cell or virus differing from parental strain in genotype (nucleotidesequence of genome)Wild-type strain: Typically refers to strain isolated from natureSelectable mutations: Those that give the mutant a growth advantage under certainconditionsNonselectable mutations: Those that usually have neither an advantage nor a disadvantageover the parent. Detection requires examining a large number of colonies and looking fordifferences (screening)Types of Mutations Silent mutation: Does not affect amino acid sequenceMissense mutation: Amino acid changed; polypeptide alteredNonsense mutation: Codon becomes stop codon; polypeptide is incompleteFrameshift mutations: Deletions or insertions that result in a shift in the reading frameReversion: Alteration in DNA that reverses the effects of a prior mutation. Point mutationsare typically reversibleMutagenesis Mutagens: chemical, physical, or biological agents that increase mutation ratesNucleotide base analogs: resemble nucleotidesChemical mutagens: induce chemical modifications, for example, alkylating agents likenitrosoguanidine20
Mutagens that cause frameshift mutations, for example, intercalating agents like acridinesNon-ionizing (UV radiation): purines and pyrimidines strongly absorb UV, and a pyrimidinedimer is one effect of UV radiationIonizing (X-rays, cosmic rays, and gamma rays): ionize water and produce free radicals,which can damage macromolecules in the cellThe Ames test makes practical use of bacterial mutations to detect for potentially hazardouschemicals. It looks for an increase in mutation rate of bacteria in the presence of suspectedmutagenGenetic Recombination Recombination: Physical exchange of DNA between genetic elementsHomologous recombination: Process that results in genetic exchange between homologousDNA from two different sourcesRecombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods ofgenetic recombination (such as molecular cloning) to bring together genetic material frommultiple sources, creating sequences that would not otherwise be found in biologicalorganismsTransformation Transformation: A genetic transfer process by which DNA is incorporated into a recipient celland brings about genetic changeDiscovered by Fredrick Griffith in the late 1920s working with Streptococcus pneumoniaeCompetent cells: cells capable of taking up DNA and being transformed. In naturallytransformable bacteria, competence is regulated. In other strains, specific procedures arenecessary to make cells competentElectricity can be used to force cells to take up DNA (electroporation)Transduction Generalized transduction: DNA derived from virtually any portion of the host genome ispackaged inside the mature virionDefective virus particle incorporates fragment of the cell’s chromosome randomlyConjunction Bacterial conjugation (mating): mechanism of genetic transfer that involves cell-to-cellcontact21
F (fertility) plasmid: a circular DNA molecule about 100 kbp long, which contains genes thatregulate DNA replication and transposable elements that allow the plasmid to integrate intothe host chromosomeSex pilus is essential for conjugation, only produced by donor cellContains tra genes that encode transfer functionsDNA synthesis is necessary for DNA transfer by conjugation: rolling circle replication22
Lecture 4: Microbial Evolution and SystematicsOrigin of Cellular Life Latest research suggest liquid water present 4.3 million years ago- conditions compatiblewith lifeFirst evidence for microbial life can be found in rocks 3.45 billion years old. Microbialformations called stromatolites commonEarly Earth was anoxic (anaerobic) and much hotter than present dayThought that life originated around hydrothermal springs on the ocean floorFirst self-replicating systems may have been RNA-based (RNA world theory), as RNA can bindsmall molecules (e.g., ATP, amino acids, proteins, other nucleotides), and has catalyticactivity; may have catalyzed its own synthesis from sugars, bases and phosphateDNA, a more stable molecule, eventually became the genetic repositoryOther Important Steps in Emergence of Cellular Life: Buildup of lipids, synthesis ofphospholipid membrane, vesicles that enclosed the cell’s biochemical and replicationmachinery, proteins in lipids23
The Evolutionary Process Prokaryotes are genetically haploid therefore genetic changes are immediately expressedMutations are changes in the nucleotide sequence of an organism’s genome which occurbecause of errors in replication, UV radiation, and other factorsPhylogeny: evolutionary history of a group of organisms inferred indirectly from nucleotidesequence dataMolecular clocks (chronometers): certain genes and proteins that are measures ofevolutionary change. Major assumptions of this approach are that nucleotide changes occurat a constant rate, are generally neutral, and are randomThe most widely used molecular clocks are small subunit ribosomal RNA (SSU rRNA) genesfound in all domains of lifeCarl Woese pioneered the use of SSU rRNA for phylogenetic studies in 1970s, and soestablished the presence of three domains of life: Bacteria, Archaea, and EukaryaComparative rRNA sequencing is a routine procedure that involves the amplification of thegene encoding SSU rRNA, sequencing of the amplified gene, and analysis of sequence inreference to other sequencesPhylogenetic Tree Graphic illustration of the relationships among sequencesBranches define the order of descent and ancestry of the nodesBranch length represents the number of changes that have occurred along that branchThe universal phylogenetic tree based on SSU rRNA genes is a genealogy of all life on Earth24
Microbial Phylogeny Domain Bacteria contains at least 80 major evolutionary groups (phyla)Many groups defined from environmental sequences alone —i.e., there are no culturedrepresentativesMany groups are phenotypically diverse - physiology and phylogeny not necessarily linkedSignature sequences: short oligonucleotides unique to certain groups of organismsPhylogenetic Methods A hybridization probe is a fragment of DNA or RNA of variable length (usually 100-1000bases long) which is used in DNA or RNA samples to detect the presence of nucleotidesequences (the DNA target) that are complementary to the sequence in the probeCan be labeled with fluorescent tags and hybridized to rRNA in ribosomes within cellsRibotyping (DNA Fingerprint) is a method of identifying microbes from analysis of DNAfragments generated from restriction enzyme digestion of genes encoding SSU rRNABy performing a Gel electrophoresis with the digested samples, the fragments can bevisualised as lines on the gel, where larger fragments are close to the start of the gel, andsmaller fragments further down. After blotting onto a matrix and probing, these lines form aunique pattern for each species and can be used to identify the origin of the DNA25
Microbial Taxonomy Taxonomy: The science of identification, classification, and nomenclature (naming organisms)Systematics: The study of the diversity of organisms and their relationshipsBacterial taxonomy incorporates multiple methods for identification and description of newspecies: phenotypic analysis, genotypic analysis, phylogenetic analysisPhenotypic Analysis Examines the morphological, metabolic, physiological, and chemical characters of the cellMorphology: Gram reaction, physical structure ,spores, cell size and shapeMotility: flagellar, amoeboid, gliding, swarmingMetabolism:- chemoorganotroph, chemolithtroph, phototrophPhysiology: pH, temperature, salt, response to oxygenChemistry: cell lipid and wall chemistryOther traits: pigments, serotype, antibiotic sensitivityGenotypic Analysis DNA–DNA hybridization: genomes of two organisms are hybridized to examine proportion ofsimilarities in their gene sequences. Provides rough index of similarity between twoorganisms, especially useful for differentiating very similar organismsDNA profiling: Several methods can be used to generate DNA fragment patterns for analysisof genotypic similarity among strains, including ribotyping, which focuses on a single gene,and repetitive extragenic palindromic PCR (rep-PCR) and amplified fragment lengthpolymorphism (AFLP), which focus on MANY genes located randomly throughout genomeMultilocus sequence typing: characterizes isolates of microbial species using the DNAsequences of internal fragments of multiple housekeeping genes. Approximately 450-500 bpinternal fragments of each gene are used, as these can be accurately sequenced on bothstrands using an automated DNA sequencer. Has sufficient resolving power to distinguishbetween very closely related strainsGC ratios: Percentage of guanine plus cytosine in an organism’s genomic DNA. Vary from 20to 80% among Bacteria and Archaea. Generally accepted that if GC ratios of two strainsdiffer by 5% they are unlikely to be closely related26
Species in Microbiology No universally accepted concept of species for prokaryotes; biological species concept(interbreeding population)not meaningful as prokayotes are haploid and do not undergosexual reproductionCurrent definition of prokaryotic species relates to a collection of strains sharing a highdegree of similarity in several independent traits27
Most important traits include: 70% or greater DNA–DNA hybridization and 97% or greater16S rRNA gene sequence identityPhylogenetic analysis: 16S rRNA gene sequences are useful in taxonomy; serve as “goldstandard” for the identification and description of new species. It is proposed that abacterium should be considered a new species if its 16S rRNA gene sequence differs by morethan 3% from any named strain, and a new genus if it differs by more than 5%The lack of divergence of the 16S rRNA gene limits its effectiveness in discriminatingbetween bacteria at the species level; thus, a multigene approach can be usedMultigene sequence analysis uses complete sequences and comparisons are made usingcladistic methodsEcotype: population of cells that share a particular resource. Different ecotypes can co-existin a habitatNew genetic capabilities can also arise by horizontal gene transfer; the extent amongbacteria is variable, and can be between speciesClassification and Nomenclature Nearly 7,000 species of Bacteria and Archaea are presently known – based primarily on 16srRNA gene sequencingProkaryotes are given descriptive genus names and species epithets following the binomialsystem of nomenclature used throughout biology: e.g. Staphylococcus aureus andMicrococcus luteusThe genus name is always written with a capital letter and species name with a small letter.Both are in italics, with latin roots frequently usedNote that Salmonella Typhimurium is not true species, so have a second capital letter28
Assignment of names for species and higher groups of prokaryotes is regulated by theInternational Code of Nomenclature of BacteriaFormal recognition of a new prokaryotic species requires deposition of a sample of theorganism in two culture collections, and official publication of the new species name anddescription in the International Journal of Systematic and Evolutionary MicrobiologyClassification Techniques Taxonomists efforts in classifying organisms according to their ancestry has resulted in lessemphasis on comparison of physically and chemical traitsHowever in routine testing laboratories phenotypic characteristics are still frequently usedto identify organismsPhysical characteristics: colony morphology on various media, shape and ability to react tovarious stainsBiochemical tests: fermentation of carbohydrates, utilization of specific substrates eg starch,citrate, gelatin, amino acids, production of waste products, fatty acid compositionSerological tests: antibody /antigen agglutination test distinguish between species andstrains within a species – highly specific eg E. coli O157:H7Phage typing: phages are specific for the hosts they infect – a strain is susceptible but arelated strain will be resistant. Used for typing SalmonellaNucleic acids: PCR and nucleotide sequencing, determination of GC ratio in a cellTaxonomic Keys Keys organize the above characteristics so they can be used to efficiently to identify anunknown organism via dichotomous keysA dichotomous key is a series of paired statements worded such that only one of twochoices applies to the particular microorganismIn the tests selected 90% of strains must respond yes or no to the test. Tests where theresponse is less than 90% are ancillary tests and useful supporting data but should not beused for preparation of a dichotomous key29
Lecture 5: Microbial Interactions with HumansOverview of Human–Microbial Interactions Most microorganisms are benign: few contribute to health and even fewer pose directthreats to healthThe human microbiome (or human microbiota) is the aggregate of microorganisms, amicrobiome that resides on the surface and in deep layers of skin (including in mammaryglands), in the saliva and oral mucosa, in the conjunctiva, and in the gastrointestinal tractsMicrobial colonization in the human body begins shortly after birth, and average adultspossess 10 times more microbial cells than human cellsPathogenicity: the ability of a parasite to inflict damage on the hostVirulence: measure of pathogenicityOpportunistic pathogen: causes disease only in the absence of normal host resistanceInfection: situation in which a microorganism is established and growing in a host, whetheror not the host is harmedDisease: damage or injury to the host that impairs host functionInfections frequently begin at sites in the animal’s mucous membranesMicroflora of the Skin The skin is generally a dry, acid environment that does not support the growth of mostmicroorganismsThe total number of bacteria on an average human has been estimated at 1012 (1 trillion).Most are found in the superficial layers of the epidermis and the upper parts of hair folliclesSkin flora is usually non-pathogenic, and either commensals (are not harmful to their host)or mutualistic (offer a benefit). The benefits bacteria can offer include preventing transientpathogenic organisms from colonizing the skin surface, either by competing for nutrients,secreting chemicals against them, or stimulating the skin's immune systemComposition is influenced by environmental factors (e.g., weather) and host factors (e.g.,age, personal hygiene)Microfl
Microbiology Subject Notes Lecture 1: Introduction to Microbiology Prokaryotes vs Eukaryotes . 2 Infectious Disease Biofilms A biofilm is any group of microorganisms in which cells stick to each other on a surface These adherent cells are frequently embedded within a self-produced matrix of extracellular