Transcription
A gentle introduction to graph neural networksSeongok Ryu, Department of Chemistry @ KAIST
Motivation Graph Neural Networks Applications of Graph Neural Networks
Motivation
Non-Euclidean data structureSuccessful deep learning architectures1. Convolutional neural networks
Non-Euclidean data structureSuccessful deep learning architectures1. Convolutional neural networks
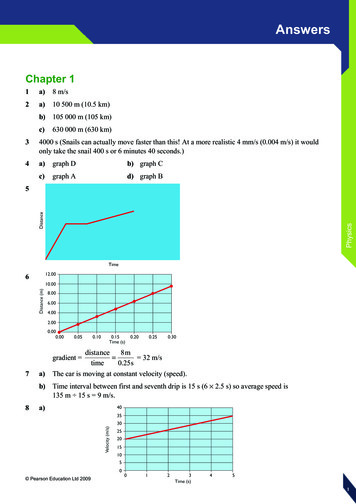
Non-Euclidean data structureSuccessful deep learning architectures1. Convolutional neural 7b852
Non-Euclidean data structureSuccessful deep learning architectures1. Convolutional neural networksLee, Jason, Kyunghyun Cho, and Thomas Hofmann."Fully character-level neural machine translation without explicit segmentation." arXiv preprint arXiv:1610.03017 (2016).
Non-Euclidean data structureSuccessful deep learning architectures2. Recurrent neural network
Non-Euclidean data structureSuccessful deep learning architectures2. Recurrent neural network
Non-Euclidean data structureSuccessful deep learning architectures2. Recurrent neural networkSentiment analysis
Non-Euclidean data structureSuccessful deep learning architectures2. Recurrent neural networkNeural machine translation
Non-Euclidean data structureData structures of shown examples are regular.Image – values on pixels (grids)Sentence – sequential structure
Non-Euclidean data structureHOWEVER, there are lots of irregular data structure, Social Graph(Facebook, Wikipedia)3D MeshAll you need is GRAPH!Molecular Graph
Graph structure𝑮𝒓𝒂𝒑𝒉 𝑮(𝑿, 𝑨)X : Node, Vertex- Individual person in a social network- Atoms in a moleculeRepresent elements of a system
Graph structure𝑮𝒓𝒂𝒑𝒉 𝑮(𝑿, 𝑨)A : Adjacency matrix- Edges of a graph- Connectivity, RelationshipRepresent relationship or interaction between elements of the system
Graph structureMore detail, Battaglia, Peter W., et al."Relational inductive biases, deep learning, and graph networks." arXiv preprint arXiv:1806.01261 (2018).
Graph structureAriana Grande 25 years old Female American Singer Node featuresDonald Trump 72 years old Male American President, business man Vladimir Putin 65 years old Male Russian President
Graph structureEdge featuresAromatic bondDouble bondSingle bond
Learning relation and interactionWhat can we do with graph neural networks?Battaglia, Peter W., et al."Relational inductive biases, deep learning, and graph networks." arXiv preprint arXiv:1806.01261 (2018).
Learning relation and interactionWhat can we do with graph neural networks? Node classification Link prediction Node2Vec, Subgraph2Vec, Graph2Vec: Embedding node/substructure/graph structure to a vector Learning physics law from data And you can do more amazing things with GNN!
Graph neural networks
Overall architecture of graph neural networks Updating node states- Graph Convolutional Network (GCN)- Graph Attention Network (GAT)- Gated Graph Neural Network (GGNN) Readout : permutation invariance on changing node orders Graph Auto-Encoders Practical issues- Skip connection- Inception- Dropout
Principles of graph neural networkWeights using in updating hidden states of fully-connected Net, CNN and RNNBattaglia, Peter W., et al."Relational inductive biases, deep learning, and graph networks." arXiv preprint arXiv:1806.01261 (2018).
Overall neural network structure– case of supervised learning(𝟎)Input node features, 𝑯𝒊: Raw node information(𝑳)Final node states, 𝑯𝒊: How the NN recognizes the nodesGraph features, Z
Principles of graph neural networkUpdates in a graph neural network Edge update : relationship or interactions, sometimes called as ‘message passing’ex) the forces of spring Node update : aggregates the edge updates and used in the node updateex) the forces acting on the ball Global update : an update for the global attributeex) the net forces and total energy of the physical systemBattaglia, Peter W., et al."Relational inductive biases, deep learning, and graph networks." arXiv preprint arXiv:1806.01261 (2018).
Principles of graph neural networkWeights using in updating hidden states of GNNSharing weights for all nodes in graph,(𝒍)but nodes are differently updated by reflecting individual node features, 𝑯𝒋
GCN : Graph Convolutional Networkhttp://tkipf.github.io/Famous for variational autoencoder (VAE)Kipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 (2016)
GCN : Graph Convolutional NetworkWhat NN learns(𝒍 𝟏)𝑿𝒊 𝝈((𝒍)(𝒍)(𝒍)𝒋 [𝒊 𝒌,𝒊 𝒌] 𝑾𝒋 𝑿𝒋 𝒃𝒋 )
GCN : Graph Convolutional Network2134(𝒍 𝟏)𝑯𝟐𝒍 𝝈 𝑯𝟏 𝑾𝒍𝒍 𝑯𝟐 𝑾𝒍𝒍 𝑯𝟑 𝑾𝒍𝒍 𝑯𝟒 𝑾𝒍
GCN : Graph Convolutional NetworkWhat NN learns(𝒍 𝟏)𝑯𝒊𝒍 𝝈𝑯𝒋 𝑾𝒋 𝑵 𝒊𝒍𝒍 𝝈 𝑨𝑯𝒋 𝑾𝒍
GCN : Graph Convolutional Network Classification of nodes of citation networks and a knowledge graph 𝐿 𝑙 𝑦𝐿𝐹𝑓 1 𝑌𝑙𝑓ln 𝑍𝑙𝑓Kipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 (2016)
GAT : Graph Attention Network Attention revisitsWhat NN learnsGCN :(𝒍 𝟏)𝑯𝒊𝒍 𝝈𝑯𝒋 𝑾𝒋 𝑵 𝒊𝒍𝒍 𝝈 𝑨𝑯𝒋 𝑾𝒍
GAT : Graph Attention Network Attention revisitsWhat NN learns: Convolution weight and attention coefficientGAT :(𝒍 𝟏)𝑯𝒊(𝒍) 𝝈𝒍𝜶𝒊𝒋 𝑯𝒋 𝑾𝒋 𝑵 𝒊𝒍Velickovic, Petar, et al."Graph attention networks." arXiv preprint arXiv:1710.10903 (2017).
GAT : Graph Attention Network Attention mechanism in natural language -neural-machine-translation-gpus-part-3/figure6 sample translations-2/
GAT : Graph Attention Network Attention mechanism in natural language and-memory-in-deep-learning-and-nlp/
GAT : Graph Attention Network Attention mechanism in natural language processing𝒂𝒕 𝑠 𝑎𝑙𝑖𝑔𝑛 𝒉𝒕 , 𝒉𝒔 𝑠𝑜𝑓𝑡𝑚𝑎𝑥 𝑠𝑐𝑜𝑟𝑒 𝒉𝒕 , 𝒉𝒔𝛽 𝒉𝑻𝒕 𝒉𝒔𝑠𝑐𝑜𝑟𝑒(𝒉𝒕 , 𝒉𝒔 ) 𝒉𝑻𝒕 𝑾𝒂 𝒉𝒔𝒗𝑻𝒂 tanh(𝑾𝒂 [𝒉𝒕 , 𝒉𝒔 ])Luong, Minh-Thang, Hieu Pham, and Christopher D. Manning."Effective approaches to attention-based neural machine translation." arXiv preprint arXiv:1508.04025 (2015).
GAT : Graph Attention Network Attention revisitsWhat NN learns: Convolution weight and attention coefficient(𝒍 𝟏)𝑯𝒊(𝒍) 𝝈𝒍𝜶𝒊𝒋 𝑯𝒋 𝑾𝒋 𝑵 𝒊𝒍𝜶𝒊𝒋 𝒇(𝑯𝒊 𝑾, 𝑯𝒋 𝑾)Velickovic, Petar, et al."Graph attention networks." arXiv preprint arXiv:1710.10903 (2017).
GAT : Graph Attention Network Attention revisitsWhat NN learns: Convolution weight and attention coefficient(𝒍 𝟏)𝑯𝒊(𝒍) 𝝈𝒍𝜶𝒊𝒋 𝑯𝒋 𝑾𝒋 𝑵 𝒊𝒍𝜶𝒊𝒋 𝒇(𝑯𝒊 𝑾, 𝑯𝒋 𝑾) Velickovic, Petar, et al. – network analysis𝜶𝒊𝒋 𝒔𝒐𝒇𝒕𝒎𝒂𝒙 𝒆𝒊𝒋 𝒆𝒊𝒋𝒆𝒙𝒑𝒌 𝑵 𝒊𝒆𝒊𝒌 Seongok Ryu, et al. – molecular applications𝜶𝒊𝒋 𝐭𝐚𝐧𝐡𝑯𝒊 𝑾 𝑻 𝑪 𝑯𝒋 𝑾𝒆𝒊𝒋 𝑳𝒆𝒂𝒓𝒌𝒚𝑹𝒆𝑳𝑼(𝒂𝑻 𝑯𝒊 𝑾, 𝑯𝒋 𝑾 )
GAT : Graph Attention Network Multi-head attention Average(𝒍 𝟏)𝑯𝒊 𝝈𝟏𝑲𝑲(𝒍)𝒍𝜶𝒊𝒋 𝑯𝒋 𝑾𝒍𝒌 𝟏 𝒋 𝑵 𝒊 Concatenation𝑲(𝒍 𝟏)𝑯𝒊 (𝒍)𝝈𝒌 𝟏𝒍𝜶𝒊𝒋 𝑯𝒋 𝑾𝒍𝒋 𝑵 𝒊Velickovic, Petar, et al."Graph attention networks." arXiv preprint arXiv:1710.10903 (2017).
GGNN : Gated Graph Neural Network Message Passing Neural Network (MPNN) frameworkUsing previous node state and message stateto update the ith hidden node state(𝑙 1)ℎ𝑖𝑙 𝑈(ℎ𝑖 , 𝑚𝑖𝑙 1)The message state is updated by previousneighbor states and the route state, and edge states.(𝑙 1)𝑚𝑖 𝑀𝑙 1(ℎ𝑖 , ℎ𝑗 , 𝑒𝑖𝑗 )𝑗 𝑁(𝑖)Wu, Zhenqin, et al."MoleculeNet: a benchmark for molecular machine learning." Chemical science 9.2 (2018): 513-530.
GGNN : Gated Graph Neural Network Using recurrent unit for updating the node states, in this case GRU.(𝒍 𝟏)𝒉𝒊𝒍 𝑼(𝒉𝒊 , 𝒎𝒊𝒍 𝟏(𝒍 𝟏))𝒉𝒊𝒍 𝑮𝑹𝑼(𝒉𝒊 , 𝒎𝒊𝒍 𝟏)Updating rate of the temporal state(𝒍 𝟏)𝒉𝒊 𝒛𝒍 𝟏 𝒉𝒊(𝒍 𝟏) 𝟏 𝒛Temporal node state𝒍 𝟏(𝒍) 𝒉𝒊Previous node stateLi, Yujia, et al."Gated graph sequence neural networks." arXiv preprint arXiv:1511.05493 (2015).
Readout : permutation invariance on changing nodes order(𝟎)Input node features, 𝑯𝒊: Raw node information(𝑳)Final node states, 𝑯𝒊: How the NN recognizes the nodesGraph features, Z
Readout : permutation invariance on changing nodes orderMapping Images to Scene Graphs with Permutation-Invariant Structured Prediction - Scientific Figure on ResearchGate. Available eling-function-F-is fig1 323217335 [accessed 8 Sep, 2018]
Readout : permutation invariance on changing nodes order Graph feature𝑧𝐺 𝑓(𝐿)𝐻𝑖 Node-wise summation𝑧𝐺 𝜏𝑀𝐿𝑃 𝐻𝑖𝐿𝑖 𝐺 Graph gathering𝐿𝑧𝐺 𝜏𝜎 𝑀𝐿𝑃1 𝐻𝑖 , 𝐻𝑖𝑖 𝐺0 𝑀𝐿𝑃2 𝐻𝑖𝐿 𝜏 : ReLU activation 𝜎 : sigmoid activationGilmer, Justin, et al."Neural message passing for quantum chemistry." arXiv preprint arXiv:1704.01212 (2017).
Graph Auto-Encoders (GAE) Clustering Link prediction Matrix completion and recommendationKipf, Thomas N., and Max Welling."Variational graph auto-encoders." arXiv preprint arXiv:1611.07308 (2016).https://github.com/tkipf/gae
Graph Auto-Encoders (GAE)Input graph, 𝑮(𝑿, 𝑨)EncoderNode features, 𝑞(𝒁 𝑿, 𝑨)DecoderReconstructedAdjacency matrix, 𝐴 𝝈(𝒁𝒁𝑻 )Kipf, Thomas N., and Max Welling."Variational graph auto-encoders." arXiv preprint arXiv:1611.07308 (2016).https://github.com/tkipf/gae
Graph Auto-Encoders (GAE)Encoder - Inference modelInput graph, 𝑮(𝑿, 𝑨) Two-layer GCN𝐺𝐶𝑁 𝑿, 𝑨 𝑨𝑅𝑒𝐿𝑈 𝑨𝑿𝑾𝟎 𝑾𝟏12 12 𝑨 𝑫 𝑨𝑫: symmetrically normalized adjacency matrix Variational Inference𝑞(𝒛𝒊 𝑿, 𝑨)𝑖 1Node features, 𝑞(𝒁 𝑿, 𝑨)Decoder𝑁𝑞 𝒁 𝑿, 𝑨 Encoder𝑞 𝒛𝒊 𝑿, 𝑨 𝑁 𝒛𝒊 𝝁𝒊 , 𝑑𝑖𝑎𝑔 𝝈𝟐𝒊ReconstructedAdjacency matrix, 𝐴 𝝈(𝒁𝒁𝑻 )Kipf, Thomas N., and Max Welling."Variational graph auto-encoders." arXiv preprint arXiv:1611.07308 (2016).https://github.com/tkipf/gae
Graph Auto-Encoders (GAE)Decoder - Generative model Inner product between latent vectors𝑁𝑁𝑝 𝑨𝒁 𝑝(𝑨𝒊𝒋 𝒛𝒊 , 𝒛𝒋 )𝑝 𝑨𝒊𝒋 𝒛𝒊 , 𝒛𝒋 𝜎 𝒛𝑇𝒊 𝒛𝒋𝑖 1 𝑗 1Input graph, 𝑮(𝑿, 𝑨)EncoderNode features, 𝑞(𝒁 𝑿, 𝑨)𝐴𝑖𝑗 : the elements of (reconstructed) A𝜎 : sigmoid activation𝑨 𝝈(𝒁𝒁𝑻 )with 𝒁 𝑮𝑪𝑵(𝑿, 𝑨) LearningDecoderReconstructedAdjacency matrix, 𝑨 𝝈(𝒁𝒁𝑻 )ℒ 𝔼𝑞(𝒁 𝑿,𝑨) log 𝑝(𝑨 𝒁) KL 𝑞 𝒁 𝑿, 𝑨 (𝑝(𝒁)Reconstruction lossKL-divergenceKipf, Thomas N., and Max Welling."Variational graph auto-encoders." arXiv preprint arXiv:1611.07308 (2016).https://github.com/tkipf/gae
Practical Issues: InceptionGoogLeNet – Winner of 2014 ImageNet 1
Practical Issues: InceptionInception guide-to-deep-network-architectures-65fdc477db41
Practical Issues: Inception(𝟏)𝑯𝒊𝟎 𝝈 𝑨𝑯𝒋 𝑾(𝟎)(𝟐)𝑯𝒊𝟏 𝝈 𝑨𝑯𝒋 𝑾(𝟏)
Practical Issues: Inception Make network wider Avoid vanishing gradient
Practical Issues: Skip-connectionResNet – Winner of 2015 ImageNet ChallengeGoing Deeeeeeeeper!
Practical Issues: Skip-connectionResNet – Winner of 2015 ImageNet Challenge(𝑙 1)𝑦 𝐻𝑖(𝑙) e-guide-to-deep-network-architectures-65fdc477db41
Practical Issues: DropoutDropout rate (𝑝) 0.25# parameters: 4x4 16# parameters: 4x4 16# parameters: 16x0.75 12# parameters: 16x0.75x0.75 9For a dense network, the dropout of hidden state reduces the number of parametersfrom 𝑁𝑤 to 𝑁𝑤 1 𝑝2
Practical Issues: DropoutHidden states in a graph neural networkGraph Conv. :(𝑙 1)𝐻𝑖 𝐴𝑯(𝒍) 𝑊Node 1Node 2Node 3 Age284036 SexMFF NationalityKoreanAmericanFrench JobStudentMedicalDoctorPolitician ��𝟒
Practical Issues: DropoutHidden states in a graph neural networkNode 1Node 2Node 3 Age284036 ctor(𝒍)𝑯𝟏(𝒍)𝑯𝟐Node 1Node 2Node 3 Age284036 SexMFF French NationalityKoreanAmericanFrench Politician JobStudentMedicalDoctorPolitician (𝒍)𝑯𝟑Mask individual ���)𝑯𝟑(𝒍)𝑯𝟒Mask information (features)of node statesAnd many other options are possible. The proper method depends on your task.
Applications of graph neural networks
Network Analysis1. Node classification2. Link prediction3. Matrix completion Molecular Applications1. Neural molecular fingerprint2. Quantitative Structure-Property Relationship (QSPR)3. Molecular generative model Interacting physical system
Network analysis1. Node classification – karate club networkKarate club graph, colors denote communities obtainedvia modularity-based clustering (Brandes et al., 2008). All figures and descriptions are taken from Thomas N. Kipf’s blog. Watch video on his blog.GCN embedding (with random weights)for nodes in the karate club network.Kipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 etworks/
Network analysis1. Node classification Good node features Good node classification resultsKipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 etworks/
Network analysis1. Node classification Semi-supervised learning – low label rate Citation network – Citeseer, Cora, Pubmed / Bipartite graph - NELLKipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 etworks/
Network analysis1. Node classification Outperforms classical machine learning methodsKipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 etworks/
Network analysis1. Node classification Spectral graph convolution𝐻(𝑙 1) 𝜎 𝐷 1/2 𝐴𝐷 1/2 𝐻 𝑙 𝑊 (𝑙)𝐴 𝐴 𝐼𝑁 , 𝐷𝑖𝑖 𝑗 𝐴𝑖𝑗 Spectral graph filtering𝑔𝜃 𝑥 𝑈𝑔𝜃′Λ𝑈𝑇 𝑥𝑈 : the matrix of eigenvectors of the normalized graph Laplacian𝐿 𝐼𝑁 11 2 2𝐷 𝐴𝐷 𝑈Λ𝑈 𝑇𝐾𝜃𝑘′ 𝑇𝑘 (Λ)𝑔𝜃 ′ Λ Polynomial approximation (In this case, Chebyshev polynomial)𝑘 0Kipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 etworks/
Network analysis1. Node classification Spectral graph convolution𝐻(𝑙 1) 𝜎 𝐷 1/2 𝐴𝐷 1/2 𝐻 𝑙 𝑊 (𝑙)𝐴 𝐴 𝐼𝑁 , 𝐷𝑖𝑖 𝑗 𝐴𝑖𝑗 Spectral graph filtering𝐾𝑔𝜃 𝑥 𝜃0′ 𝑥 𝜃1′ 𝐿 𝐼𝑁 𝑥 𝜃0′ 𝑥 𝜃1′ 𝐷 1/2 𝐴𝐷 1/2 𝑥𝜃𝑘′ 𝑇𝑘 𝐿 𝑥𝑔𝜃 𝑥 Linear approx.𝑘 0𝑔𝜃 𝑥 𝜃 𝐷 1/2 𝐴𝐷 1/2 𝑥Use a single parameter 𝜃 𝜃0′ 𝜃1′𝑔𝜃 𝑥 𝜃 𝐼𝑁 𝐷 1/2 𝐴𝐷 1/2 𝑥Renormalization trick𝐼𝑁 𝐷 1/2 𝐴𝐷 1/2 𝐷 1/2 𝐴𝐷 1/2Kipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 etworks/
Network analysis1. Node classification Spectral graph convolution𝐻(𝑙 1) 𝜎 𝐷 1/2 𝐴𝐷 1/2 𝐻 𝑙 𝑊 (𝑙)𝐴 𝐴 𝐼𝑁 , 𝐷𝑖𝑖 𝑗 𝐴𝑖𝑗Kipf, Thomas N., and Max Welling."Semi-supervised classification with graph convolutional networks." arXiv preprint arXiv:1609.02907 etworks/
Network analysis2. Link prediction Clustering Link prediction Matrix completion and recommendationKipf, Thomas N., and Max Welling."Variational graph auto-encoders." arXiv preprint arXiv:1611.07308 (2016).https://github.com/tkipf/gae
Network analysis2. Link predictionInput graph, 𝑮(𝑿, 𝑨)EncoderNode features, 𝑞(𝒁 𝑿, 𝑨)DecoderReconstructedAdjacency matrix, 𝐴 𝝈(𝒁𝒁𝑻 )Kipf, Thomas N., and Max Welling."Variational graph auto-encoders." arXiv preprint arXiv:1611.07308 (2016).https://github.com/tkipf/gae
Network analysis2. Link prediction Trained on an incomplete version of {Cora, Citeseer, Pubmed} datasets where parts of the citation links(edges) have been removed, while all node features are kept. Form validation and test sets from previously removed edges and the same number of randomlysampled pairs of unconnected nodes (non-edges).Kipf, Thomas N., and Max Welling."Variational graph auto-encoders." arXiv preprint arXiv:1611.07308 (2016).https://github.com/tkipf/gae
Network analysis3. Matrix completion Matrix completion Can be applied for recommending systemBerg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Network analysis3. Matrix completion A rating matrix 𝑀 of shape 𝑁𝑢 𝑁𝑣𝑁𝑢 : the number of users, 𝑁𝑣 : the number of items User 𝑖 rated item 𝑖, or the rating is unobserved (𝑀𝑖𝑗 0). Matrix completion problem or recommendation a link prediction problem on a bipartite user-item interaction graph.Input graph : 𝑮(𝓦, 𝓔, 𝓡) 𝓦 𝓤 𝓥 : user nodes 𝒖𝐢 𝓤, with 𝑖 {1, , 𝑁𝑢 } and item nodes 𝒗𝒋 𝓥, with 𝑗 1, , 𝑁𝑣 𝒖𝒊 , 𝑟, 𝒗𝒋 𝓔 represent rating levels, such as 𝑟 1, , 𝑅 𝓡.Berg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Network analysis3. Matrix completion GAE for the link prediction taskTake as input an 𝑁 𝐷 feature matrix, 𝑿𝑁 𝐸 node embedding matrix, 𝒁 𝒛𝑻𝟏 , , 𝒛𝑻𝑵𝑻𝒁 𝑓(𝑿, 𝑨)A graph adjacency matrix, 𝑨𝑨 𝑔(𝒁)which takes pairs of node embeddings 𝒛𝒊 , 𝒛𝒋 and predicts respective entries 𝑨𝒊𝒋Berg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Network analysis3. Matrix completion GAE for the bipartite recommender graphs, 𝑮(𝓦, 𝓔, 𝓡)Encoder𝑈, 𝑉 𝑓(𝑋, 𝑀1 , , 𝑀𝑅 ), where 𝑀𝑟 0,1𝑁𝑢 𝑁𝑣is the adjacency matrix associated with rating type 𝑟 ℛ𝑈, 𝑉 : matrices of user and item embeddings with shape 𝑁𝑢 𝐸 and 𝑁𝑣 𝐸, respectively.Decoder𝑀 𝑔(𝑈, 𝑉), rating matrix 𝑀 of shape 𝑁𝑢 𝑁𝑣𝐺(𝑿, 𝑨)𝐺(𝓦, 𝓔, 𝓡)𝒁 𝑓(𝑿, 𝑨)𝑨 𝑔(𝒁)𝑼, 𝑽 𝑓(𝑿, 𝑴𝟏 , , 𝑴𝑹 )𝑴 𝑔(𝑼, 𝑽)GAE for the link prediction taskGAE for the bipartite recommenderBerg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Network analysis3. Matrix completion GAE for the bipartite recommender graphs, 𝑮(𝓦, 𝓔, 𝓡)𝐺(𝓦, 𝓔, 𝓡)𝑼, 𝑽 𝑓(𝑿, 𝑴𝟏 , , 𝑴𝑹 )𝑴 𝑔(𝑼, 𝑽)Berg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Network analysis3. Matrix completionEncoder𝑢𝑖 𝜎 𝑊ℎ𝑖 : the final user embeddingℎ𝑖 𝜎 𝑎𝑐𝑐𝑢𝑚𝜇𝑗 𝑖,1 , ,𝑗 𝒩𝑖 ,1𝜇𝑗 𝑖,𝑟 1𝑊𝑥𝑐𝑖𝑗 𝑟 𝑗𝜇𝑗 𝑖,𝑅: intermediate node state𝑗 𝒩𝑖 ,𝑅: Message function from item 𝑗 to user i𝑐𝑖𝑗 𝒩𝑖 𝒩𝑗Berg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Network analysis3. Matrix completionDecoder𝑝 𝑀𝑖𝑗 𝑟 𝑒𝑢𝑖𝑇 𝑄𝑟 𝑣𝑗𝑇𝑢𝑖 𝑄𝑠 𝑣𝑗𝑠 𝑅 𝑒𝑀𝑖𝑗 𝑔 𝑢𝑖 , 𝑣𝑗 𝔼𝑝𝑀𝑖𝑗 𝑟: probability that rating 𝑀𝑖𝑗 is 𝑟𝑟 𝑟 𝑝(𝑀𝑖𝑗 𝑟): expected rating𝑟 𝑅Loss function𝑅ℒ 𝐼 𝑟 𝑀𝑖𝑗 log 𝑝 𝑀𝑖𝑗 𝑟𝐼 𝑘 𝑙 1, when 𝑘 𝑙 and 0 otherwise𝑖,𝑗 𝑟 1Berg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Network analysis3. Matrix completionBerg, Rianne van den, Thomas N. Kipf, and Max Welling."Graph convolutional matrix completion." arXiv preprint arXiv:1706.02263 (2017).
Molecular applications: Which kinds of datasets exist? Bioactive molecules with drug-like properties 1,828,820 compounds https://www.ebi.ac.uk/chembldb/ Drugs and targets FDA approved, investigational, experimental, 7,713 (all drugs), 4,115 (targets), https://drugbank.ca/ Drugs and targets FDA approved, investigational, experimental, 7,713 (all drugs), 4,115 (targets), https://drugbank.ca/
Molecular applications: Which kinds of datasets exist?Tox21 Data Challenge (@ Kaggle) 12 types of toxicity Molecule species (represented with SMILES)and toxicity labels are given But too small to train a DL sp
Molecular applications1. Neural molecular fingerprintHash function have been used to generate molecular fingerprints.* Molecular fingerprint : a vector representation of molecular ii/
Molecular applications1. Neural molecular fingerprintSuch molecular fingerprints can be easily obtained by open source packages, e.g.) RDKit.http://kehang.github.io/basic roperty-prediction/
Molecular applications1. Neural molecular fingerprintIn recent days, neural fingerprints generated by graph convolutional network is widely used formore accurate molecular property predictions.http://kehang.github.io/basic roperty-prediction/
Molecular applications1. Neural molecular fingerprintDuvenaud, David K., et al."Convolutional networks on graphs for learning molecular fingerprints." Advances in neural information processing systems. 2015.
Molecular applications1. Neural molecular fingerprintDuvenaud, David K., et al."Convolutional networks on graphs for learning molecular fingerprints." Advances in neural information processing systems. 2015.
Molecular applications1. Neural molecular fingerprintDuvenaud, David K., et al."Convolutional networks on graphs for learning molecular fingerprints." Advances in neural information processing systems. 2015.
Molecular applications1. Neural molecular fingerprintDuvenaud, David K., et al."Convolutional networks on graphs for learning molecular fingerprints." Advances in neural information processing systems. 2015.
Molecular applications2. Quantitative Structure-Property Relationships (QSPR)Ryu, Seongok, Jaechang Lim, and Woo Youn Kim."Deeply learning molecular structure-property relationships using graph attention neural network." arXiv preprint arXiv:1805.10988 (2018).
Molecular applications2. Quantitative Structure-Property Relationships (QSPR)Ryu, Seongok, Jaechang Lim, and Woo Youn Kim."Deeply learning molecular structure-property relationships using graph attention neural network." arXiv preprint arXiv:1805.10988 (2018).
Molecular applications2. Quantitative Structure-Property Relationships (QSPR)Learning solubility of moleculesFinal node statesThe neural network recognizesseveral functional groups differentlyRyu, Seongok, Jaechang Lim, and Woo Youn Kim."Deeply learning molecular structure-property relationships using graph attention neural network." arXiv preprint arXiv:1805.10988 (2018).
Molecular applications2. Quantitative Structure-Property Relationships (QSPR)Learning photovoltaic efficiency (QM phenomena)Final node statesInterestingly, The NN also can differentiate nodesaccording to the quantum mechanical characteristics.Ryu, Seongok, Jaechang Lim, and Woo Youn Kim."Deeply learning molecular structure-property relationships using graph attention neural network." arXiv preprint arXiv:1805.10988 (2018).
Molecular applications2. Quantitative Structure-Property Relationships (QSPR)Learning solubility of moleculesAscendingorderGraph features𝒁𝒊 𝒁𝒋𝟐Similar molecules are located closelyin the graph latent space
Molecular applications3. Molecular generative modelMotivation : de novo molecular design Chemical space is too huge: only 108 molecules have beensynthesized as potential drug candidates,whereas it is estimated that there are1023 to 1060 molecules. Limitation of virtual screening
Molecular applications3. Molecular generative modelMotivation : de novo molecular designMolecule GraphSimplified Molecule Line-Entry System(SMILES)Molecules can be represented as strings according to defined rules
Molecular applications3. Molecular generative modelMotivation : de novo molecular designMany SMILES-based generative models existSegler, Marwin HS, et al. "Generating focused molecule libraries for drug discoverywith recurrent neural networks." ACS central science 4.1 (2017): 120-131.Gómez-Bombarelli, Rafael, et al. "Automatic chemical design using a data-drivencontinuous representation of molecules." ACS central science 4.2 (2018): 268-276.
Molecular applications3. Molecular generative modelMotivation : de novo molecular designSMILES representation also has a fatal problem thatsmall changes of structure can lead to quite different expressions. Difficult to reflect topological information of molecules
Molecular applications3. Molecular generative modelLiteratures Li, Y., Vinyals, O., Dyer, C., Pascanu, R., & Battaglia, P. (2018). Learning deep generative models of graphs.arXiv preprint arXiv:1803.03324. Jin, Wengong, Regina Barzilay, and Tommi Jaakkola. "Junction Tree Variational Autoencoder for MolecularGraph Generation." arXiv preprint arXiv:1802.04364 (2018). Constrained Graph Variational Autoencoders for Molecule Design." arXiv preprint arXiv:1805.09076(2018). "GraphVAE: Towards Generation of Small Graphs Using Variational Autoencoders." arXiv preprintarXiv:1802.03480 (2018). De Cao, Nicola, and Thomas Kipf. "MolGAN: An implicit generative model for small molecular graphs."arXiv preprint arXiv:1805.11973 (2018).
Molecular applications3. Molecular generative modelLi, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative model1. Sample whether to add a new node of a particular type or terminate : if a node type is chosen2. Add a node of this type to the graph3. Check if any further edges are needed to connect the new node to the existing graph4. If yes, select a node in the graph and add an edge connecting the new to the selected node.Li, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative model Determine that add a node or notLi, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative model If a node is added, determine that add edges between current node and other nodes.Li, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative modelLi, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative modelGraph propagation process : readout all node states and generating a graph feature𝒉𝑮𝑽𝒉𝑮 or𝑣 𝒱𝒉′𝒗 𝑓𝑛 𝒂𝒗 , 𝒉𝒗𝒈𝑮𝒗 𝒉𝐺𝑣𝒉𝑮 𝒈𝑮𝒗 𝜎 𝑔𝑚 𝒉𝒗𝑣 𝒱 𝑣 𝒱𝒂𝒗 𝑓𝑒 𝒉𝒖 , 𝒉𝒗 , 𝒙𝒖,𝒗𝑢,𝑣 ℰ 𝑣 𝒱Li, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative modelAdd node : a step to decide whether or not to add a node𝒇𝒂𝒅𝒅𝒏𝒐𝒅𝒆 𝑮 softmax 𝑓𝑎𝑛 𝒉𝑮(𝑻)If “yes”The new node vectors 𝒉𝑽 are carried over to the next stepLi, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative modelAdd edge : a step to add an edge to the new node(𝑻)𝑓𝑎𝑑𝑑𝑒𝑑𝑔𝑒 𝐺, 𝑣 𝜎 𝑓𝑎𝑒 𝒉𝑮 , 𝒉𝒗: the probability of adding an edge to the newly created node 𝑣.𝑻If “yes”(𝑻)𝑓𝑛𝑜𝑑𝑒𝑠 𝐺, 𝑣 softmax 𝑓𝑠 𝒉𝒖 , 𝒉𝒗: Score for each node toconnect the edgesLi, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative modelObjective function𝑝(𝐺) : marginal likelihood𝑝 𝐺 permutation𝑝(𝐺, 𝜋) 𝔼𝑞𝜋 𝒫(𝐺)𝜋 𝐺𝑝(𝐺, 𝜋)𝑞 𝜋 𝐺Following all possible permutations is intractable samples from data : 𝑞 𝜋 𝐺 𝑝𝑑𝑎𝑡𝑎 (𝜋 𝐺)𝔼𝑝𝑑𝑎𝑡𝑎 (𝐺,𝜋) log 𝑝(𝐺, 𝜋) 𝔼𝑝𝑑𝑎𝑡𝑎(𝐺) 𝔼𝑝𝑑𝑎𝑡𝑎𝜋 𝐺log 𝑝(𝐺, 𝜋)Li, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative modelLi, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Molecular applications3. Molecular generative modelLi, Yujia, et al."Learning deep generative models of graphs." arXiv preprint arXiv:1803.03324 (2018).
Interacting physical system Interacting systems Nodes : particles, Edges : (physical) interaction between particle pairs Latent code : the underlying interaction graphKipf, Thomas, et al."Neural relational inference for interacting systems." arXiv preprint arXiv:1802.04687 (2018).
Interacting physical systemInput graph : 𝓖 𝓥, 𝓔 with vertices 𝑣 𝓥 and edges 𝑒 (𝑣, 𝑣 ′ ) ℰ𝑣 𝑒 𝒉𝒍(𝒊,𝒋) 𝑓𝑒𝑙 𝒉𝒍𝒊 , 𝒉𝒍𝒋 , 𝒙𝒊,𝒋𝒉𝒍 𝒊,𝒋 , 𝒙𝒋𝑒 𝑣 𝒉𝒍 𝟏 𝑓𝑣𝑙𝒋𝒙𝒊 : initial node features, 𝒙𝒊,𝒋: initial edge features𝑖 𝒩𝑗Kipf, Thomas, et al."Neural relational inference for interacting systems." arXiv preprint arXiv:1802.04687 (2018).
Interacting physical systemKipf, Thomas, et al."Neural relational inference for interacting systems." arXiv preprint arXiv:1802.04687 (2018).
Interacting physical systemEncoder𝒉𝟏(𝒊,𝒋) 𝑓𝑒1 𝒉𝟏𝒊 , 𝒉𝟏𝒋𝒉𝟐𝒋 𝑓𝑣1𝑖 𝑗𝒉𝟏(𝒊,𝒋)𝒉𝟐(𝒊,𝒋) 𝑓𝑒2 𝒉𝟐𝒊 , 𝒉𝟐𝒋𝒉𝟏𝒋 𝑓𝑒𝑚𝑏 (𝒙𝒋 )𝑞𝜙 𝑧𝑖𝑗 𝑥 softmax 𝑓𝑒𝑛𝑐,𝜙 𝑥𝑖𝑗,1:𝐾2 softmax ℎ(𝑖,𝑗)Kipf, Thomas, et al."Neural relational inference for interacting systems." arXiv preprint arXiv:1802.04687 (2018).
Interacting physical system𝑀𝑆𝐺𝑗𝑡 Decoder𝒉𝒕 𝟏 𝐺𝑅𝑈𝒋𝒉𝒕(𝒊,𝒋) 𝑧𝑖𝑗,𝑘 𝑓𝑒𝑘 𝒉𝒕𝒊 , 𝒉𝒕𝒋𝑘𝑖 𝑗𝒉(𝒊,𝒋)𝒕𝑀𝑆𝐺𝑗𝑡 , 𝒙𝒕𝒋 , 𝒉𝒋𝝁𝒕 𝟏 𝒙𝒕𝒋 𝑓𝑜𝑢𝑡 𝒉𝒋𝒋𝒕𝒕 𝟏𝑝 𝒙𝒕 𝟏 𝒙𝒕 , 𝒛 𝒩 𝝁𝒕 𝟏 , 𝜎 2 𝐈Kipf, Thomas, et al."Neural relational inference for interacting systems." arXiv preprint arXiv:1802.04687 (2018).
Interacting physical systemℒ 𝔼 𝑞𝜙𝑝𝜃 𝒙 𝒛 𝑝 𝒛 𝑇𝑡 1 𝑝𝜃𝒊 𝒋 𝑝𝜃𝒙𝒕 𝟏 𝒙𝒕 , , 𝒙𝟏 𝒛 𝒛 𝒙𝑇𝑡 1 𝑝𝜃log 𝑝𝜃 𝒙 𝒛 KL 𝑞𝜙 𝒛 𝒙 𝑝(𝒛)𝒙𝒕 𝟏 𝒙𝒕 𝒛 , since the dynamics is Markovian.𝒛𝒊𝒋 , the prior is a factorized uniform distribution over edge typesKipf, Thomas, et al."Neural relational inference for interacting systems." arXiv preprint arXiv:1802.04687 (2018).
Interacting physical systemKipf, Thomas, et al."Neural relational inference for interacting systems." arXiv preprint arXiv:1802.04687 (2018).
Interacting physical systemKipf, Thomas, et
Principles of graph neural network Updates in a graph neural network Edge update : relationship or interactions, sometimes called as 'message passing' ex) the forces of spring Node update : aggregates the edge updates and used in the node update ex) the forces acting on the ball Global update : an update for the global attribute ex) the net forces and total energy of the .