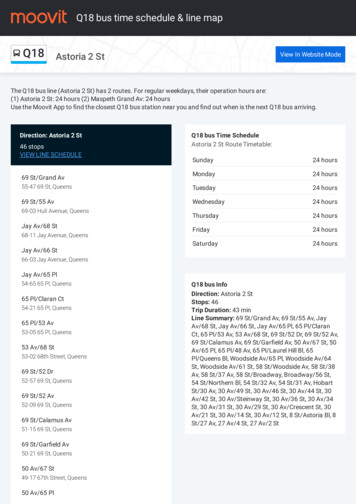
Transcription
Veterinary Research ForumVol: 2, No: 3, September, 2011, 147 - 155Review ArticleNutrigenomics and its Applications in Animal ScienceVaishali Ghormade1Ankur Khare2*R.P.S.Baghel31Department of Animal Breeding and Genetics, Faculty of Veterinary Science & A.H.,Jabalpur (M.P.)-462008, India2Department of Animal Nutrition, Faculty of Veterinary Science & A.H.,Jabalpur (M.P.)-462008, India3Dean, Faculty of Veterinary Science and A.H.Jabalpur (M.P.)-462008, IndiaReceived: 01 July 2011, Accepted: 25 August 2011AbstractNutrigenomics applies genomic technologies to study how nutrients affect expression of genes.With the advent of the post genomic era and with the use of functional genomic tools, the newstrategies for evaluating the effects of nutrition on production efficiency and nutrient utilization arebecoming available. Nutrigenomics plays an efficient role in various fields of animal health likenutrition, production, reproduction, disease process etc. Nutrigenomic approaches will enhanceresearchers‟ abilities to maintain animal health, optimize animal performance and improve milk andmeat quality.Key words: Nutrigenomics, Genotype, Nutritional management*Corresponding author:Ankur Khare, DVM, PhDDepartment of Animal Nutrition, Faculty of Veterinary Science & A.H., Jabalpur (M.P.)-462008, IndiaE-mail address: ankur khare@yahoo.com
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 155IntroductionIn recent years, nutritional research hasmoved from classical epidemiology andphysiology to molecular biology andgenetics.Followingthistrend,nutrigenomics has emerged as a novel andmultidisciplinary research field innutritional science that aims to elucidatehow diet can influence animal health.1It is already well known that bioactivefood compounds can interact with genesaffecting transcription factors, proteinexpression and metabolite production. Theapplication of modern molecular malnutrition,asopportunities to integrate the informationcoded in the genome to applied animalnutrition because; production is the goal ofanimal nutritionists. “Genomics” is thestudy of the functions and interactions ofall genes in the genome; “nutrigenomics”applies genomic technologies to studyhow nutrients affect expression of genes.The study of how genes and gene productsinteract with dietary chemicals to alterphenotype and, conversely, how genes andtheir products metabolize nutrients iscallednutritionalgenomicsor“Nutrigenomics”.2 It is expected thatnutritional genomics will be a key area innutritional research over the next decade 3and that nutrigenomic studies will be veryuseful for elucidating the roles of foodcomponents in obesity,4 coronary heartdiseases 5 and cancer prevention.6 From anutrigenomic perspective, nutrients aredietary signals, detected by the cellularsensor system, that influence gene andprotein expression and, ics is the two way interactionof individual genetic and specific dietarycomponents that may alter geneexpression and, ultimately, transcription ofan enzyme or other protein. Further, whennutrigenomics is combined with metabolicinformation (metabolomics), whole animal148assessment may be achieved and providesthe opportunity for corrective interventionvia specific nutrients and othercompounds.8With the advent of the post genomic eraand with the use of functional genomictools, the new strategies for evaluating theeffects of nutrition on productionefficiency and nutrient utilization arebecoming available. New tools thatexamine the regulation of metabolism atthe level of gene expression are being usedto examine nutritional effects of dietarycomponents and nutritional strategies.These tools will allow much more rapidevaluations of the relationship betweendiet and their key biological functions andalso promise to provide methods fordetecting fine differences in nutrientquality, and may eventually be used toevaluate not only supplementation andmanagement practices in livestockproduction systems, but also thenutritional value of feed ingredients.9From the research perspective, toexplore the effect of dietary componentson the genome, the crucial stages abolomics.Application of these modern researchtools, known as “omics” technologies,should yield new knowledge on the courseof molecular processes in animalorganisms and a more precise evaluationof the biological properties of feeds.Applications of Nutrigenomics inAnimal Science1) To develop animal feed/foodmatching to its genotype. The goal ofnutrigenomics or nutritional genomics isto develop foods and feeds that can bematched to genotypes of animals tobenefit health and enhance normalphysiological processes. Using gene chipsthat contain the genetic code of animal,researchers can measure the effects ofcertain nutritional supplements, and howthey alter the gene interactions of thebody.
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 1552) To select nutrients fine-tuned withgenes of animal. Nutrigenomics is notaltering the genetics of an animal nor togenetically modify the animal rather weare altering the activity of genes switchingon good genes and keeping bad onesswitched down. Through nutrigenomicswe are carefully selecting nutrients forfine-tuning genes and DNA present inevery cell and every tissue of an animal.For example, keeping stress responsegenes switched down with proper nutritionso that the animal is healthier, moreproductive.3) To understand role of ase) of animal. Geneexpression studies will allow for theidentification of pathways and candidategenes responsible for economicallyimportant traits. Dietary manipulationsand nutritional strategies are key tools forinfluencing ruminant production. There isa usual belief that nutrition and geneticmakeup both strongly influence thereproductive performance of milkinganimals. This is particularly importantduring the transition period and earlylactation, when the animal is particularlysensitive to nutritional imbalances.Nutrigenomics and nutritional genomicsare providing new tools that can be used tomore clearly understand how nutritionalmanagement can be applied to addressdisease, performance and productivity inanimals. In the changing scenario ofruminant‟s nutrition the objective ofnutrigenomics is to study the effects ofdiet on changes in gene expression orregulatory processes that may beassociated with various biologicalprocesses related with animal health andproduction. In studies of steers undernutritional restriction due to intake of poorquality feeds, expression of specific genesassociatedwithproteinturnover,cytoskeletal remodeling, and metabolichomeostasis was clearly influenced bydiet.10 These studies provide application ofnutrigenomics to resolve the molecularmarkers important in nutrition research.There is vary scares information abouteffect of diet on expression of genesrelated to productive or reproductive traitsof livestock, it may be possible to begin tounderstand the importance of therelationship between individual nutrientsand the regulation of gene expression. Tounderstand this concept of nutrigenomics astudy of diet induced gene expression isdiscovered in which selenium deficiencyshown to alter protein synthesis attranscriptional level.11 It leads to adverseeffect like enhancement of stress throughup-regulation of specific gene expressionand signaling pathway. On the other handgenes responsible for detoxificationmechanism and protection from oxidativedamagewerehampered,theseconsequences ultimately leads to alterationof phenotypic expression of relatedsymptoms of selenium deficiency. Fromthe above example it is apparent thatpossibly nutrigenomics can be used toidentify the specific markers to manipulategene expression through use of nutrientsor their combinations so as to improveproductive as well as overall animalperformance. Nutrigenomics will be apath-breaking tool through identificationof pathways and candidate genesresponsible for dietary induced diseasesand ultimately reduction in productionlosses due to these diseases in animals.The discoveries of gene markers related toeconomically important traits like milk,meat, wool production etc whoseexpression can be improved by dietaryregimens is a need of today‟snutrigenomic research, which will help forsustainable livestock production.4) Nutrient-Gene interaction. The diethas long been regarded as a complexmixture of natural substances that suppliesboth the energy and building blocks todevelop and sustain the organism.However, nutrients have a variety ofbiological activities. Some nutrients havebeen found to act, as radical scavengersknown as antioxidants and as such are149
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 155involved in protection against diseases.Other nutrients have shown to be potentsignaling molecules and act as nutritionalhormones.7 Some of the plant secondarymetabolites also known as photochemicalact as a modulator of animal health andproduction.5) To understanding the aging processin animals. A nutrigenomic approach canbe applied to understanding the agingprocess in companion animals. Healthyadult animals given the same foods can bestudied to identify the gene expression andbiochemical differences characteristic ofthe aging process. Foods for senioranimals can then be rationally designedand evaluated for their ability to modifygene expression profiles in animals tomore closely reflect those found in healthyadult animals, which has the potential toimprove health and quality of life. Inaddition, canine and feline nutrigenomicstudies may provide evidence thatnutrigenomics can improve health andquality of life for humans.6) Nutrigenomics and immune system.The concept underlying nutrigenomics isthat nutrition is the key element of healthmaintenance, particularly for the immunesystem, so that an optimum level ofnutrition will ensure optimum animalhealth. A deficiency of an essentialnutrient will eventually affect the body‟sperformance. The immune system isparticularly sensitive to deficiencies, andonce the immune system is compromised,negative consequences follows.There is a defined relationship betweenproduction and immune status of animals.Higher the production, more sensitive isthe immune system of animal. Twodecades ago, the main aim of animalnutritionists was to design ration so toavoid deficiencies. Deficiency is now rarein modern livestock production systems.So we can now move to the next stagerather than merely preventing deficiency,we can strive to actually meet the animal‟sexact requirements from its diet, in orderthat it can meet its genetic potential.1508) Nutrigenomics and diseases.Essential nutrients and other bioactivefood components can serve as importantregulators of gene expression patterns.Macronutrients, vitamins, minerals, andvarious phytochemicals can modify genetranscription and translation, which canalter biological responses such asmetabolism,cellgrowth,anddifferentiation, all of which are importantin the disease process. Genome widemonitoring of gene expression using DNAmicroarrays allows the simultaneousassessment of the transcription ofthousands of genes and of their relativeexpression between normal cells anddiseased cells or before and after exposureto different dietary components. Thisinformation should assist in the discoveryof new biomarkers for disease diagnosisand prognosis prediction and of newtherapeutic tools.Many diseases and disorders are relatedto suboptimal nutrition in terms venttoxicconcentrations of certain food compounds.There are multietioetiological diseaseswhich are due to interaction of differentnutrients along with several genes12 Dueto remarkable diversity in all living beingsdifferences in food digestion, nutrientabsorption, metabolism, and excretionhave been observed and genetic diseasesin these processes have been reported. Thefunctional integrity of gene is mainlydepends on metabolic signals that thenucleus receives from internal factors, e.g.hormones, and external factors, e.g.nutrients, which are among the mostinfluential of environmental stimuli.Genomes evolve in response to manytypes of environmental stimuli, includingnutrition. Therefore, the expression ofgenetic information can be highlyregulated by, nutrients, micronutrients,and photochemicals found in food.139) Nutrigenomics and reproduction.The science of nutrigenomics has begun touse information obtained from basic
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 155studies of the genome to evaluate theeffects of diet and nutrient managementschemes on gene expression. Preliminarystudies have shown the value of suchtechniques and suggest that it will bepossible to use specific gene expressionpatterns to evaluate the effects of nutritionon key metabolic processes relating toreproductive performance. While theeffects of nutrition on fertility are onlypartiallyunderstood,modernnutrigenomics will undoubtedly play a keyrole in developing strategies foraddressing some of the limitations inreproductive performance.Currently, oligo based and cDNAmicroarray techniques make it possible tounderstand many of the factors controllingthe regulation of gene transcription andglobally evaluate gene expression profilesby looking at the relative abundance ofgene-specific mRNA in tissues. Thesetechniques provide an unprecedentedamount of information and are only nowbeing used to examine key istics in cattle. They also promiseto provide a tremendous amount of newinformation that can be used to understandand diagnose key issues that limitreproductive he sequencing of the genomes has ledto the development of a whole newscientific methodology. These new areasof scientific study usually include the'omics' suffix. The technical developmentshave given us novel tools enabling highthroughput genome wide approaches.These tools form the basis of the NA),proteomics(protein), metabolomics (metabolites) andsystems biology (integrating all of these),with bioinformatics enabling the storage,integrationandanalysisoftheoverwhelmingly complex data setproduced.Nutrigenomics aims to determine theinfluence of common dietary ingredientson the genome, and attempts to relate theresultingdifferentphenotypestodifferences in the cellular and/or geneticresponse of the biological system.15 Morepractically, nutrigenomics describes theuse of functional genomic tools to probe abiological system following a nutritionalstimulus that will permit an increasedunderstandingofhownutritionalmolecules affect metabolic pathways andhomeostatic control. Nutrigenetics, on theother hand, aims to understand how thegenetic makeup of an individualcoordinates their response to diet, and thusconsidersunderlyinggeneticpolymorphisms. It embodies the science ofidentifying and characterizing genevariants associated with differentialresponses to nutrients, and relating thisvariation to disease states. Therefore, bothdisciplines aim to unravel diet/genomeinteractions; however, their approachesand immediate goals are distinct.Nutrigenomics will unravel the optimaldiet from within a series of nutritionalalternatives, whereas, nutrigenetics willyield critically important information thatwill assist clinicians in identifying theoptimal diet for a given individual, i.e.personalized nutrition.RoleoftranscriptomicsinnutrigenomicsThe transcriptome is the complete set ofRNA that can be produced from thegenome. Transcriptomics is the study ofthe transcriptome, i.e. gene expression atthe level of the mRNA.Using either cDNA or oligonucleotidemicroarray technology, it describes theapproach in which gene expression(mRNA) is analysed in a biologicalsample at a given time under specificconditions. It is the most widely used ofthe “omics” technologies. Regulation ofthe rate of transcription of genes by foodcomponents represents an intriguing sitefor regulation of an individual‟sphenotype.16 A host of essential nutrients151
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 155and other bioactive food components canserve as important regulators of iption and translation, which canalter biological responses such asmetabolism,cellgrowth,anddifferentiation.The aim of transcriptomics is todetermine the level of all or a selectedsubset of genes based on the amount ofRNA present in tissue samples.Transcriptomics is concerned with theexpression of genes in animals In preciseexperiments conducted on animals, thescope of investigation is usually restricted,for example to assess the influence ofdietary components on the transcript levelof selected organs, as demonstrated inTable 1.The use of a microarray containingprobes for the over 8000 genes present inthe liver of rats demonstrated that about 33% of the genes of rats fed a soya proteindiet differed from those of casein fedanimals.17 Significant differences wereobserved in the gene cluster concernedwith lipid metabolism, and in the generelated to energy metabolism, transcriptionfactor, and anti-oxidization enzymes. Insimilar experiments carried out byTachibana et al., (2005),18 compared withcasein, soya protein changed theexpression of 120 genes involved in lipidmetabolism, antioxidant activity andenergy metabolism. Endo et al., (2002)also reported that various dietary proteinsources resulted in a difference inexpression of about 281 genes in rat liver,suggesting a nutritional function forprotein components.19In dairy industry, an effective utilizationof microarray technology was beneficial tostudy mammary gland tissues (milkproduction and udder health), musclegrowth and development and myogenesisprocess (beef production) and the role ofgut microflora on nutritional diet intake inruminants (health and food safety). Study152of Ron et al., (2007) has effectively beenhybridized Affymetrix microarray (MGU74v2) in identification of 249differentially expressed probe setscommon to the three experiments alongthe four developmental stages of puberty,pregnancy,20 lactation and involution. Incontext to candidate genes for milkproduction traits, a total of 82 expressedgenes were identified in mammary glandtissue with at least 3-fold expression overthe median representing all issues tested inGene atlas.In pork industries, the impact ofadvanced nutrigenomics tools has beendiscussed for the economic benefits and toimprove human nutrition and health. Inpig, nutrigenomics tools were effectivelyutilized in analysis of regulation ofmyogenesisanditsbiochemicalpathways.21 Combination of biochemicalpathway and microarray results revealedthe biological insight of porcinemyogenesis process is controlled by twodistinct waves, i.e. Notch signalingpathway and the WNT signaling pathway.Recent study on porcine microarrayexpression profiling in 16 tissues revealedthe interaction between gene and tissue fordifferentially expressed genes ome.22 Evidence from therecently published transcriptomics basednutritional studies performed in livestockspecies suggests that, with appropriatestudy design, it is feasible to applytranscriptomic methods successfully inanimal feed and nutritional research.However, newly emerged nutrigenomicstools like gene expression basedbiomarker development still poses a majorchallenge and the use of expression profile„signatures‟, rather than single genes, mayprovide an eventual solution for thisinformation should assist in the discoveryof new biomarkers for disease diagnosisand prognosis prediction and of newtherapeutic tools.
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 155Role of proteomics in nutrigenomicsProteomics is the study of all theproteins in a particular cell, tissue orcompartment.23 The major tools ofproteomics are two dimensional (2D) gelelectrophoresis and mass spectrometry(MS). Proteomic analysis was quiteeffective and useful to evaluate the effectof dietary methionine on breast-meataccretion and protein expression inskeletal muscle of broiler chickens.24 Via atandem mass spectrometer, a total of 190individual proteins were identified fromPectorali major muscle tissue; three ofthem were recognized which differeddistinctly between the treatment proteomeand could be considered as potentialbiomarkers regulated by a methioninedeficiency in broiler chickens.Role of metabolomics in nutrigenomicsMetabolomics represents the final stepin understanding the function of genes andtheir proteins. The aim of metabolomics isto determine the sum of all metabolites(other substances than DNA, RNA orprotein) in a biological system: organism,organ, tissue or cell (Müller and Kersten,2003).Techniquesemployedtoinvestigate the metabolome includenuclear magnetic resonance (NMR)spectroscopy, high performance s spectrometry (GCMS). These methods are capable ofresolving and quantifying a wide range ofcompounds in a single sample. The maincharacteristics of these new technologieswere miniaturization, automation, highthroughput and computerization.25As in the case of transcriptomics andproteomics, the scope of metabolomicanalysis is mainly restricted to theassessment of the influence of dietarycomponents on the metabolome ofselected organs or tissue in animalnutritionstudies.Inexperimentsperformed by Bertram et al. (2006)metabolomic analysis was implemented todetect the changes in the biochemicalprofiles of plasma and urine from pigs fedwith high-fibre rye bread.26 Two diets withsimilar levels of dietary fibre andmacronutrients, but with contrasting levelsof wholegrain ingredients, were preparedfrom whole rye and fed to pigs. Using anexplorative approach, the studies disclosedthe biochemical effects of a wholegraindiet on plasma betaine content andexcretion of betaine and creatinine.To select nutrients fine-tuned withgenes of animalNutrigenomics and reproductionTo develop animal feed/foodmatching to its genotypeNutrigenomics anddiseasesNutrigenomicsNutrient-Gene interactionTo understand role of nutritional managementin performance (production/disease) of animal(production/disease) of animalTo understanding the agingprocess in animalsNutrigenomics and immunesystemFig 1. Schematic overview of integration of Omics technology in animal feeding and nutritional research153
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 155Table 1: Influence of Dietary Components on The Transcript LevelOmic robes, spotted cDNAproductsNuclear magneticresonance (NMR)spectroscopy,Highperformanceliquid metry (GC-MS)Nutritional ;Individualizedfood;Functional food; DiagnosisPost transitional modification;Protein-protein interaction;Bioprocess cultivation conditions;Food quality control;Functional food;DiagnosisUnderstanding the function of genes andtheir proteins;Resolving and quantifying a wide range ofcompounds in a single sample (diet);Detect the changes in the biochemicalprofiles of plasma, urine etc from animalfed with diet prepared;To determine the sum of all metabolites(other substances than DNA, RNA orprotein) in a biological system: organism,organ, tissue or cellAnimal FeedingQuality of animal feedstuff;Quality of animal products;Optimizationofnutrient/diets requirements;Food safety assessmentConclusionReferencesNutrigenomic approaches will enhanceresearchers‟ abilities to maintain animalhealth, optimize animal performance andimprove milk and meat quality.Nutrigenomics is a rapidly emergingscience still in its beginning stages. It isuncertain whether the tools to studyprotein expression and metaboliteproduction have been developed to thepoint as to enable efficient and reliablemeasurements. Also once such researchhas been achieved, it will need to beintegrated together in order to produceresults and dietary recommendations. Allof these technologies are still in theprocess of development.1. Canas VG, Simo C, Leon C, et al.Advances in Nutrigenomics research:novel and future analytical approaches toinvestigate the biological activity ofnatural compounds and food functions. JPharm Biomed Anal 2009;51(2):290304.2. Kaput J, Ordovas JM, Ferguson L, et al.The case for strategic internationalalliances to harness nutritional genomicsfor public and personal health. Brit JNutr 2005;94:623-32.3. Trayhurn P. Nutritional genomics"Nutrigenomics". British Journal Nutrition2003;89: 1-2.4.ChadwickR.Nutrigenomics,individualism and public health. ProcNutr Soc 2004;63:161-166.5. Talmud PJ. How to identify geneenvironmentinteractionsinamultifactorial disease: CHD as anexample. Proc Nutr Soc 2004;63: 5-10.6. Davis CD, Hord NG. Nutritional“omics” technologies for elucidating therole(s) of bioactive food components inAcknowledgementI want to acknowledge our newlyformed Veterinary University ” and our Hon‟ble ViceChancellor Dr. G.P.Mishra.154
V. Ghormade et al / Veterinary Research Forum. 3(September, 2011) 147 - 155colon cancer prevention. J Nutr2005;135: 2694-2697.7. Muller M, Kersten S. Nutrigenomics:Goals and Strategies. Nat Rev Genet2003;4:315-322.8. Lundeen T. Nutrigenomics speeds(Nutrition & Health: Dairy). Feedstuffs,10.9. Dawson KA, Harrison GA. UsingNutrigenomicApproachesforUnderstanding Forage Quality andNutrient Restriction. In: 22nd AnnualSouthwest Nutrition & ManagementConference, 2007; 22-23.10. Byrne KA, Wang YH, Lehnert SA, etal Gene expression profiling of muscletissue in Brahman steers duringnutritional restriction. J Anim Sci2005;83:1-12.11. Rao L, Puschner B, Prolla TA. Geneexpression profiling of low seleniumstatus in the mouse intestine:transcriptional activation of genes linkedto DNA damage, cell cycle control andoxidative stress. J Nutr 2001;131:31753181.12. Mariman C M. Nutrigenomics andnutrigenetics: the „omics‟ revolution innutritional science. Biotechnol ApplBiochem 2006;44:119-128.13. Van Ommen B. Nutrigenomics:Exploiting systems biology in thenutrition and health arenas. Nutrition2004;20:2-8.14. Dawson KA. Nutrigenomics: Feedingthe genes for improved fertility. AnimReprod Sci 2006;96:312-222.15. Mutch, D. M., Wahli, W. &Williamson, G., 2005. Nutrigenomicsand Nutrigenetics: the emerging faces ofnutrition. FASEB J 2008;19: 1602–1616.16. Trujillo E, Davis C, Milner J.Nutrigenomics,proteomics,metabolomics, and the practice ofdietetics. J Am Diet Assoc 2006;106(3):403-13.17. Takamatsu K, Tachibana N,Matsumoto I, et al. Soy proteinfunctionality and nutrition analysis.Biofactors 2004;21:49-53.18. Tachibana N, Matsumoto I, Fukui K,et al. Intake of soy protein isolate altershepatic gene expression in rats. J AgrFood Chem 2005; 53: 4253-4257.19. Endo Y, Fu Z, Abe K, et al. Dietaryprotein quantity and quality effect rathepatic gene expression. J Nutr2002;132: 3632-3637.20. Ron M, Israeli G, Seroussi E, et al.Combining mouse mammary gland geneexpression and comparative mapping forthe identifi cation of candidate genes forQTL of milk production traits in cattle.BMC Genomics 2007;8:183-193.21. Te Pas MF, Hulsegge I, Coster A, et al.Biochemical pathways analysis ofmicroarray results: regulation ofmyogenesis in pigs. BMC Develop Biol2007;7: 66-80.22. Ferraz A, Ferraz LZ, Ojeda A, et al.Transcriptome architecture across tissuesin the pig. BMC Genomic 2008;9:173180.23. Banks RE, Dunn MJ, Hochstrasser DF,et al. Proteomics: New perspectives, newbiomedicalopportunities.Lancet2000;356: 1749-1756.24. Corzo A, Kidd MT, Dozier WA, et al.Protein expression of pectoralis majormuscle in chickens in response to dietarymethionine status. Brit J Nutr2006;95:703-708.25. Corthesy-Theulaz I, den Dunnen JT,Ferre P, et al. Nutrigenomics: Theimpact of biomics technology onnutrition research. Ann Nutr Metab2005; 49: 355-365.26. Bertram HC, Bach Knudsen KE,SerenaA,etal.NMR-basedmetabolomic studies reveal changes inthe biochemical profile of plasma andurine from pigs fed high-fibre rye bread.Brit J Nutr 2006;95: 955-962.155
addition, canine and feline nutrigenomic studies may provide evidence that nutrigenomics can improve health and quality of life for humans. 6) Nutrigenomics and immune system. The concept underl