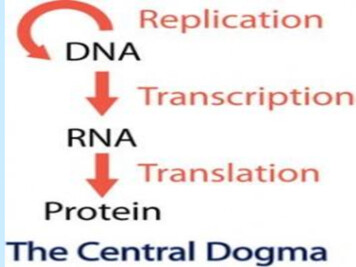
Transcription
DNA Replicationand Repair
/imgorg/cendog.gif
genetic information is passed onto the next generation ATATGCGCGCGATATATCGCGCGTATATATATATCGCGCAGParent moleculewith ch parentalstrand is atemplateEach daughterDNA moleculeconsists of oneparental and onenew strand
Overview of replicationInitiation DNA is unwound and stabilized Origins of replication: Replication bubble and replication forkPriming RNA primers bind to sections of the DNA and initiate synthesisElongation Leading strand (5’ 3’) synthesized continuously Lagging strand synthesized discontinuously then fragments are joined RNA primer replaced by DNAProofreading Mismatch repair by DNA polymerase Excision repair by nucleases
Review of DNA structure double helix each strand has a 5’phosphate end and a 3’hydroxyl end strands run antiparallelto each other A-T pairs (2 H-bonds),G-C pairs (3 H-bonds)
STEP 1Initiation at originsof replicationseparation sites on DNA strands Depend on a specific AT-rich DNA sequence– Prokaryotes – one site– Eukaryotes – multiple sites Replication bubble Replication fork Proceeds in two directions from point oforigin
The proteinsof initiation1. Helicase –unwinds doublehelix2. Single-strandbinding proteins– holds DNA apart3. Topoisomerase –relieves strain bybreaking,swiveling,rejoining strands
STEP 2Priminginitiation of DNA synthesis by RNARNA primers bind tounwound sectionsthrough the actionof primase– leading strand –only 1 primer– lagging strand –multiple primers– replaced by DNAlater
STEP 3Elongation of a new DNAstrandlengthening in the 5’ 3’ directionDNA polymerase III can only addnucleotides to the 3’ hydroxylendLeading strand- DNA pol III – adds nucleotidestowards the replication fork;- DNA pol I - replaces RNA withDNALagging strand- DNA pol III - adds Okazakifragments to free 3’ end awayfrom replication fork- DNA pol I - replaces RNA withDNA- DNA ligase – joins Okazakifragments to create acontinuous strand
STEP 4Proofreadingcorrecting errors in replicationMismatch repair DNA pol III – proofreadsnucleotides against thetemplate strandExcision repair nuclease – cuts damagedsegment DNA pol III and ligase – fill thegap leftTelomeres at 5’ ends of laggingstrands no genes, only 100 – 1000TTAGGG sequences to protectgenes telomerase catalyzeslengthening of telomeres
DNA Replication and Repair1. Summarize the central dogma in a diagram.2. Define antiparallel and semiconservative in terms ofthe structure of DNA.3. Use the following terms associated with replicationand create a flowchart showing the different stages:replication bubble and replication fork, helicase,single-strand binding proteins, RNA primer, primase,leading strand, lagging strand, DNA polymerase III,DNA polymerase I, DNA ligase, Okazaki fragments,and 5’à 3’.4. Differentiate between mismatch and excision repair.5. What are telomeres and what role do they play inprotecting the integrity of the lagging strand of theDNA?
Modelling1. By team, create a DNA strand that is at least20 nucleotide pairs long with at least onestretch that has the sequence ATATAA2. One member should be sketching the DNAstrand on the sheet provided3. Indicate the 5’ end and the 3’ end for eachstrand
Modelling4. You are modeling eukaryotic DNA. Howwould prokaryotic DNA be different?5. Use the clay to create helicase,topoisomerase, and single-strand bindingproteins.6. Show how these act in unwinding, stabilizingand holding the strands apart.
Modelling7. In real life, RNA primers are 7-10 nucleotideslong. Create two 3-nucleotide long RNAprimers that would correspond to thesequence complementary strands closest tothe two replication forks.8. Create primase using clay and use it to attachthe RNA primers to the correct sequences onthe complementary DNA strand.
DNA Replication and Repair 1. Summarize the central dogma in a diagram. 2. Define antiparallel and semiconservative in terms of the structure of DNA. 3. Use the following terms associated with replication and create a flowchart showing the different stages: replication bubble and replication fork, helicase,